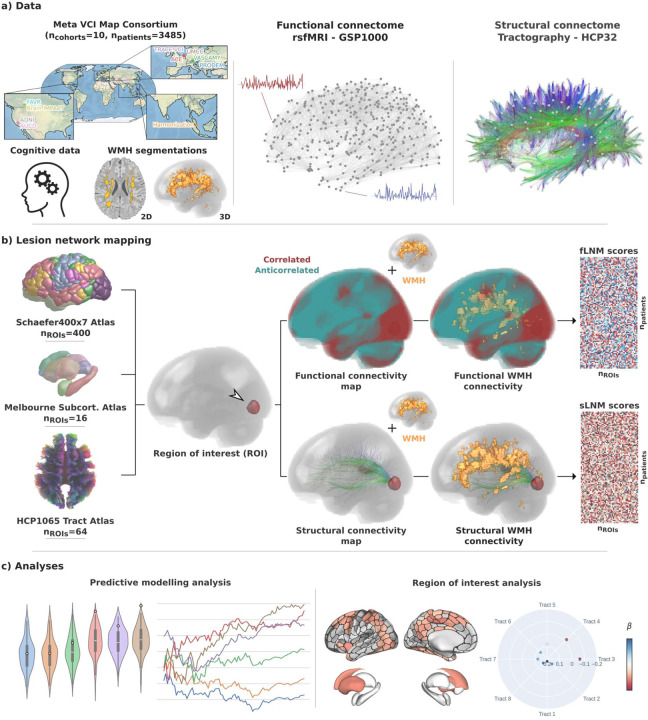
Figure 1. Methodology.
a) Data from 10 memory clinic cohorts of the Meta VCI Map Consortium were used including harmonized cognitive scores and WMH segmentations in MNI space. For functional LNM we employed the GSP1000 normative functional connectome comprising resting-state fMRI data from 1000 healthy GSP participants. For structural lesion network mapping, we used the HCP32 normative structural connectome based on diffusion-weighted imaging data from 32 healthy HCP participants, detailing the fiber bundle architecture. b) LNM was performed to quantify the functional and structural connectivity of WMH to multiple ROIs (Schaefer400×7 cortical, Melbourne Subcortical Atlas subcortical, HCP1065 white matter areas). For this, voxel-level functional and structural connectivity maps were computed for each ROI, reflecting resting-state BOLD correlations or anatomical connection strength via tractography streamlines, respectively. ROIwise LNM scores were derived by averaging voxel-level connectivity indices within the normalized WMH masks, considering only positive correlation coefficients for functional mapping. This resulted in a matrix for both fLNM and sLNM scores per ROI per patient (nROIs × npatients). The matrices shown in the figure are populated with random data only serving as a visual aid. c) The fLNM and sLNM scores across patients were used in predictive models to estimate cognitive domain scores (predictive modelling analysis) and analyzed in permutation-based general linear models to identify regions significantly influencing the cognitive domain-WMH disconnectivity relationship at the ROI level (ROI-level inferential statistics). Abbreviations: fLNM = functional lesion network mapping, GSP = Genomic Superstruct Project, HCP = Human Connectome Project, ROI = region of interest, rsfMRI = resting-state functional magnetic resonance imaging, sLNM = structural lesion network mapping, WMH = white matter hyperintensities of presumed vascular origin.