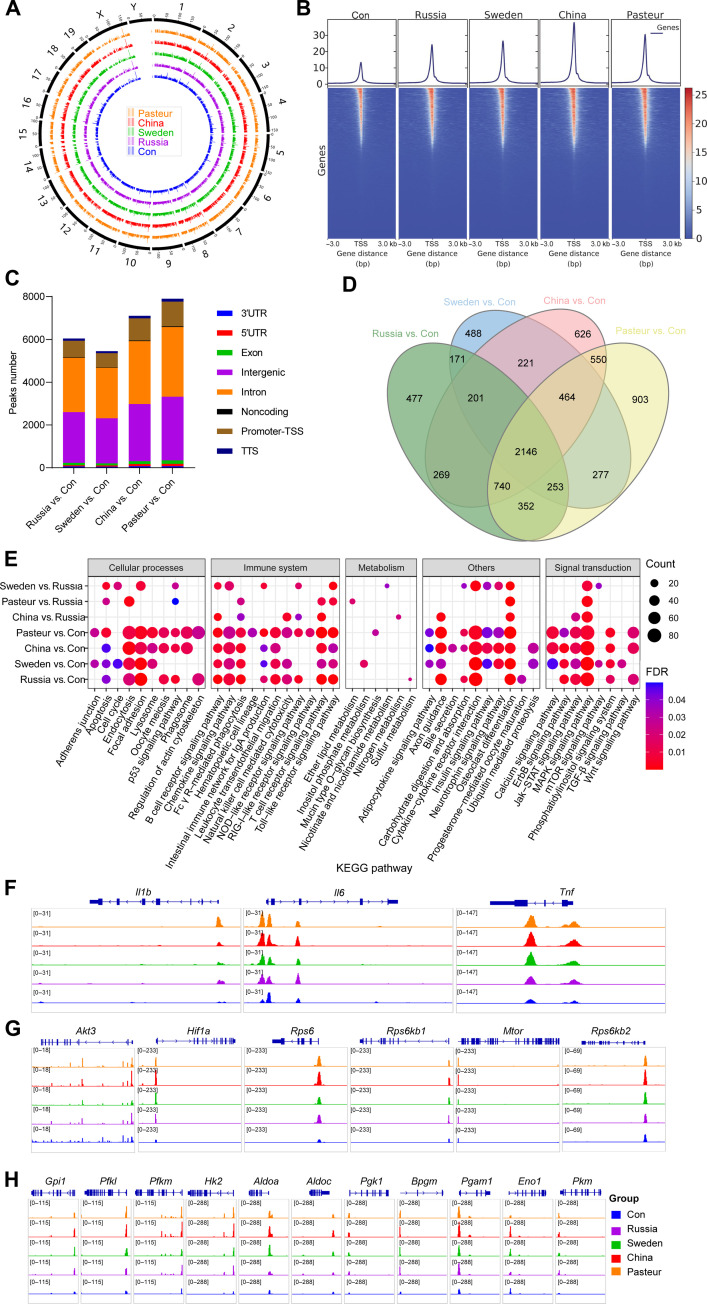
Fig. 5. The epigenetic landscape of BMDM cells after BCG training.
(A) Genome-wide chromatin accessibility of trained BMDMs. (B) Heatmaps of differential assay for transposase accessible chromatin (ATAC) peak (P < 0.05) densities at transcriptional start site (TSS) regions. bp, base pairs. (C) Differential ATAC peak distributions on gene loci. 3′UTR, untranslated region; 5′UTR, untranslated region; TTS, transcription termination site. (D) Venn diagram of genes with differential ATAC peaks. (E) Heatmap comparison between BCG-trained groups, KEGG enrichment results of genes with differential ATAC peaks [false discovery rate (FDR) < 0.05]. (F) ATAC signals of Il1b, Il6, and Tnf gene. (G) ATAC signals of genes involved in the HIF-1 signaling pathway. (H) ATAC signals of genes involved glycolysis. Data represent three independent biological duplicates.