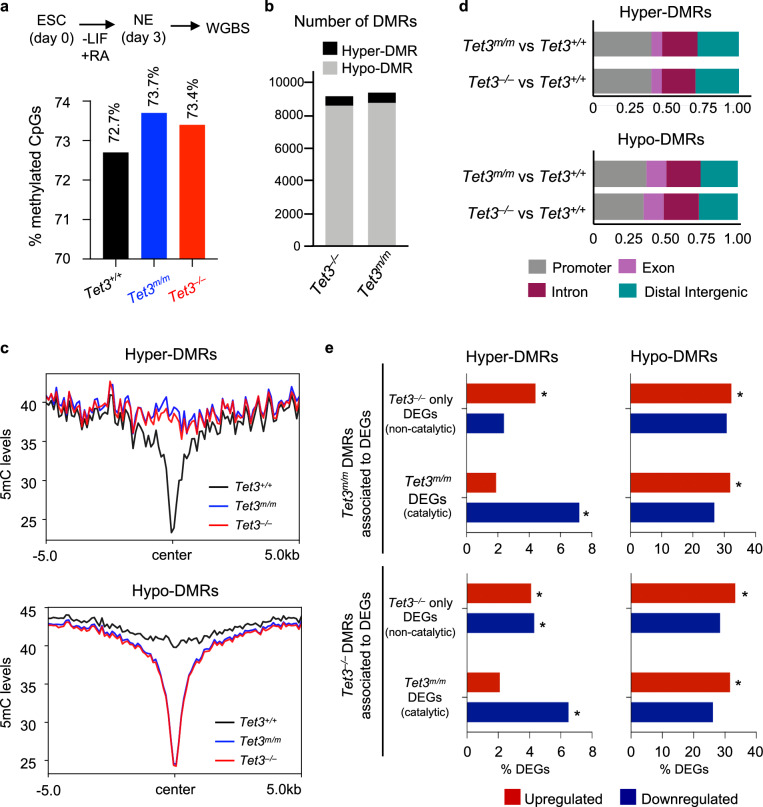
Fig. 3. Genome wide analysis of DNA methylation in Tet3m/m, and Tet3–/– NE cells by WGBS.
a Percent methylated CpG sites genome-wide in NE cells of indicated genotypes by Whole Genome Bisulfite Sequencing (WGBS). b Hyper- and hypo- differentially methylated regions (DMRs = >5 CpGs, methylation difference >20%, and FDR < 0.05) in Tet3m/m and Tet3–/– (compared to Tet3+/+) NE cells. c DNA methylation levels at all hyper-DMRs (n = 924) and all hypo-DMRs (n = 12,050) in Tet3m/m and Tet3–/– cells. d Annotation of hyper- and hypo-DMRs to genomic regions. e Percent of catalytic and non-catalytic DEGs that overlap with all hyper- and hypo-DMRs in both Tet3m/m and Tet3–/– NE cells. * Hypergeometric test p-value < 0.05.