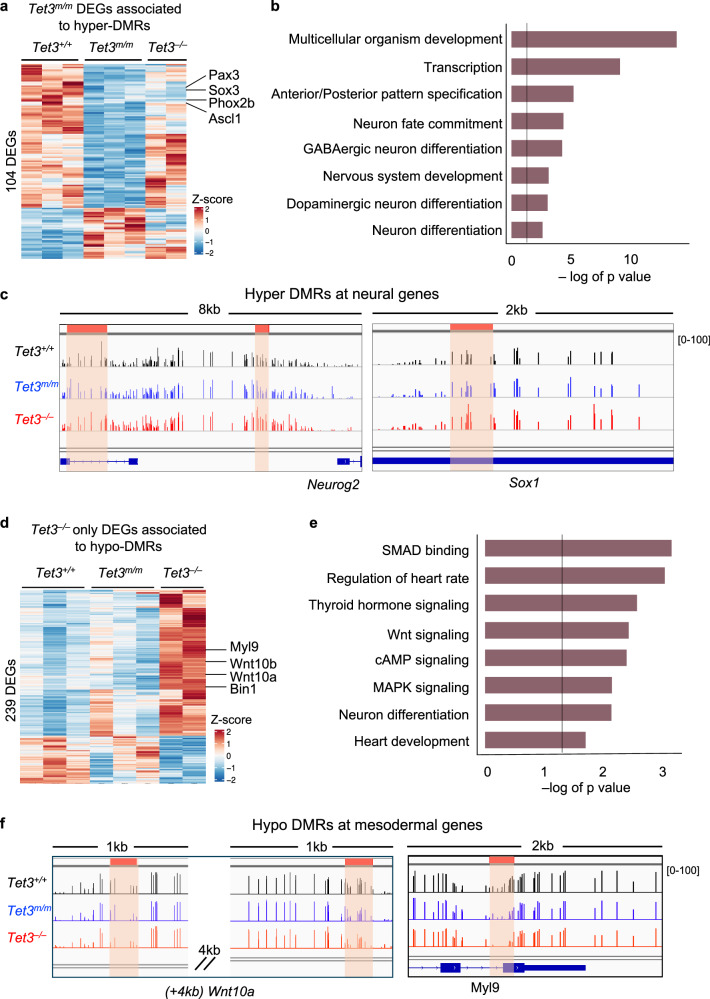
Fig. 4. Hyper- and hypo-DMRs are associated to neural and mesodermal genes, respectively.
a Heatmap of all TET3 catalytic target genes (n = 104) associated with hyper-DMRs. b GO analysis of hyper-DMR containing TET3 catalytic target genes revealing enrichment for neural terms. Line represents significance (p < 0.05). c IGV tracks of DNA methylation levels at Neurog2 and Sox1 loci (both downregulated in Tet3m/m and Tet3–/–). Hyper-DMRs are highlighted in orange. d Heatmap of all TET3 non-catalytic target genes (n = 239) associated with hypo-DMRs. e GO analysis of hypo-DMR-containing TET3 non-catalytic target genes revealing enrichment for mesodermal signaling pathways. Line represents significance (p < 0.05). f IGV tracks of DNA methylation levels at Wnt10a and Myl9 loci (both upregulated in Tet3–/– but not in Tet3m/m NEs). Hypo-DMRs are highlighted in orange.