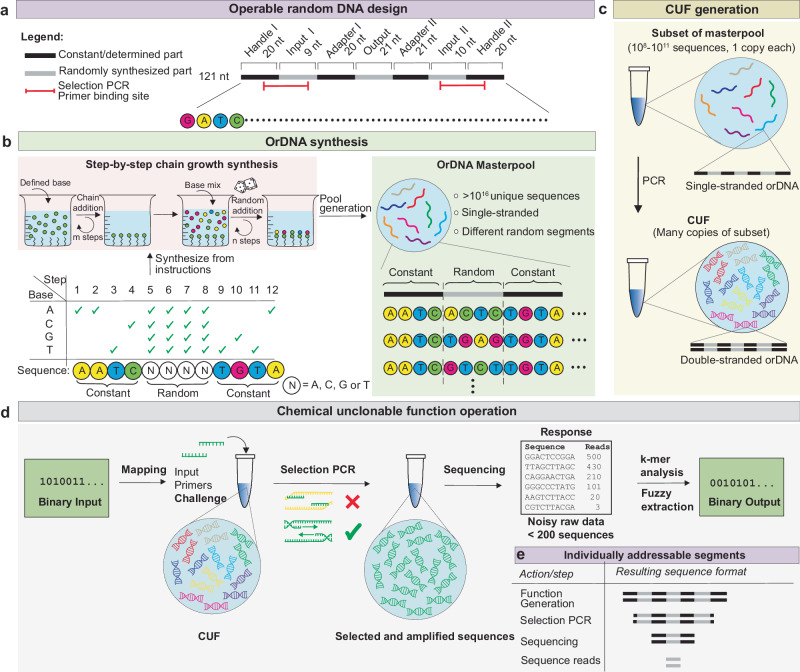
Fig. 1. Design and working principle of DNA-based chemical unclonable functions.
a Library design of the implemented operable random DNA (orDNA), containing constant handle and adapter segments that separate three random elements (two input and one output segment). The segment in red indicates the binding site of the input primers. b Step-by-step chemical synthesis of orDNA, combining random with sequence-determined synthesis, as offered by many commercial suppliers. c CUF generation procedure, amplifying a subset from the synthesized orDNA masterpool to generate multiple double-stranded copies of each sequence. d Working principle of a DNA-based one-way function. A numeric input is mapped to two primer sequences, which selectively amplify matching sequences out of a large DNA pool of a random composition. The resulting selection is sequenced, leading to a “fingerprint” characteristic for any given input, from which a binary, fixed-length output is generated using numeric data processing (k-mer analysis and fuzzy extraction). e Overview of the different orDNA segments involved in each step from function generation to readout.