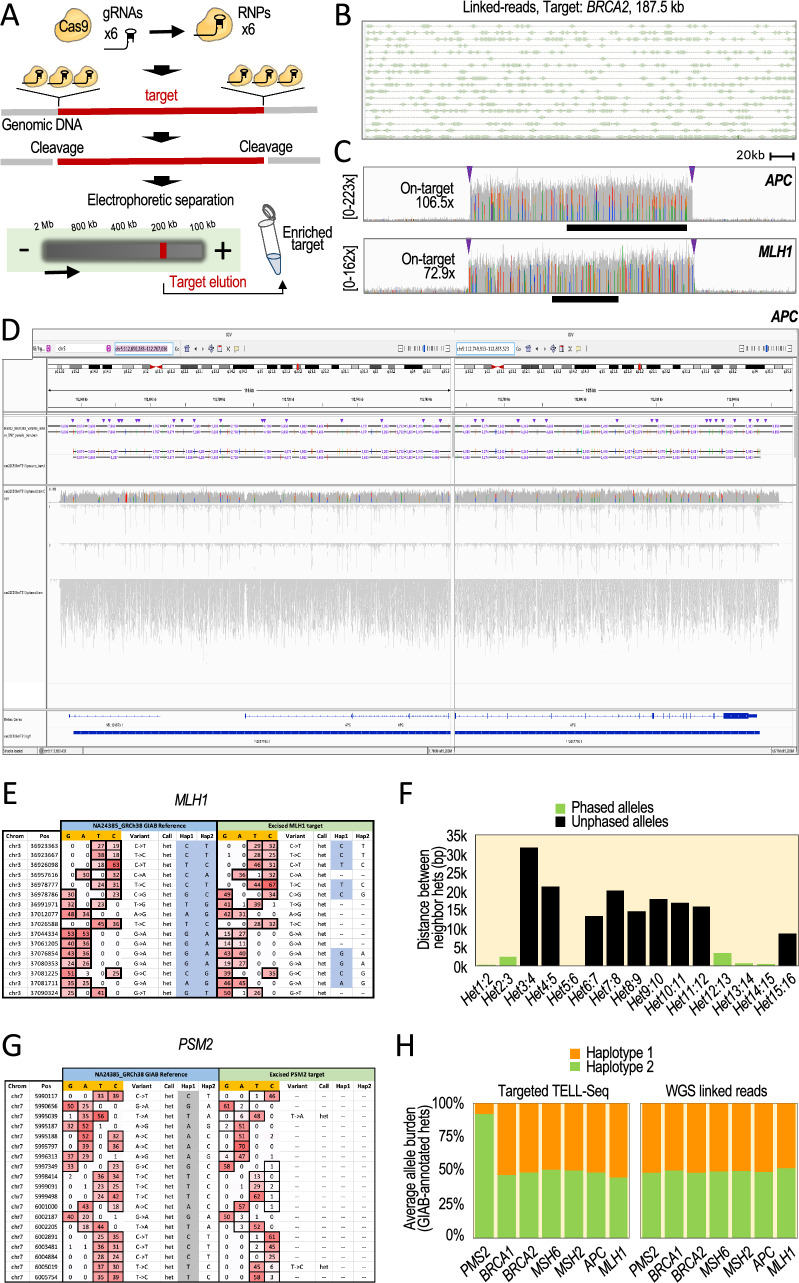
Figure 2.
Phasing 185–205 kb HLS-CATCH-enriched targets. (A) Experimental workflow. See also Supplementary Fig. S1A. (B) A representative example of linked reads obtained with the targeted TELL-Seq protocol. Shown a 186 kb region in the BRCA2 target. Short reads (boxes) with identical barcode are connected by lines (i.e., linked reads). (C) Read profiles (coverage indicated) from TELL-Seq libraries showing robust on-target recovery compared to background. Targets: 200 kb APC and MLH1. Arrowheads represent locations of a trio of gRNA binding sites on each target end. Black bars represent the APC and MLH1 genes. (D) Joint screenshots from the Integrative Genomics Viewer (IGV) Portal showing TELL-Seq results for the APC target. Tracks (from top to bottom): GIAB phased haplotypes in HG002 (benchmark); phased haplotypes (numbers indicate distances between some phased sites; coverage; inferred haplotype 1; inferred haplotype 2; unphased reads; gene annotations; and inferred phase block. (E, G) Read behaviors across representative regions in the MLH1 (E) and PMS2 (G) targets showing correct genotyping but incomplete phasing (squared cells) and heterozygosity loss (incomplete genotyping and phasing) affecting Hap2 (squared cells), respectively. Left columns represent GIAB data (NA24385), right columns represent TELL-Seq data. Counts per positions are indicated by nucleotide G, A, T, C columns). Variants calls are also indicated (Variant columns). (F) Distances between the annotated neighbor heterozygous sites. Green columns represent phased sites. Black columns represent unphased sites. (H) Average read counts (allele burden) by target relative to the total read count. Sites selected based on GIAB annotated haplotypes.