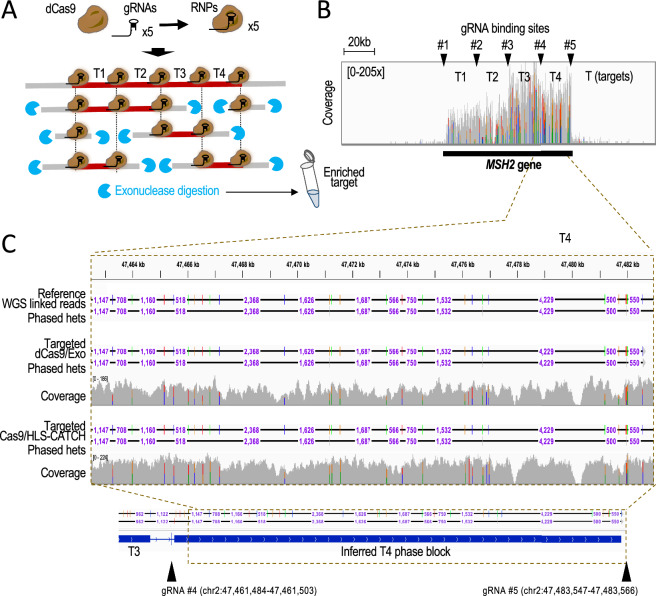
Figure 3.
TELL-Seq-mediated phasing of four adjacent 20 kb targets enriched with dCaBagE, a modified CaBagE method that leverages dCas9 to protect end and internal sites of the target. (A) Experimental workflow. The entire (80 kb) or sub-targets (20, 40, or 60 kb) can be individually phased based on the quality of the input genomic DNA and the presence of free DNA ends, which serve as sites for exonuclease initiation of degradation. The protected DNA (target) is represented in dark red; unprotected DNA (non-target) is represented in grey. (B) Coverage across the MSH2 locus (black bar indicates the position of the MSH2 gene). This profile supports high on-target recovery over background for all sub-targets (T1–T4) but higher for T3–T4 than for T1–T2. Arrowheads indicate gRNA binding sites (#1–#5). (C) Screenshot from the IGV Portal showing phased data and coverage using targeted TELL-Seq with dCas9/exonuclease or HLS-CATCH/Cas9 as DNA pre-enrichment methods. Shown on top HG002 WGS linked-read phased data (benchmark). Phased data includes distances between phased sites (we note that distances are not shown when not allowed at the selected resolution on IGV). Bottom tracks show phased data and inferred phase blocks with targeted TELL-Seq using dCaBagE. Arrowheads indicate gRNA binding sites.