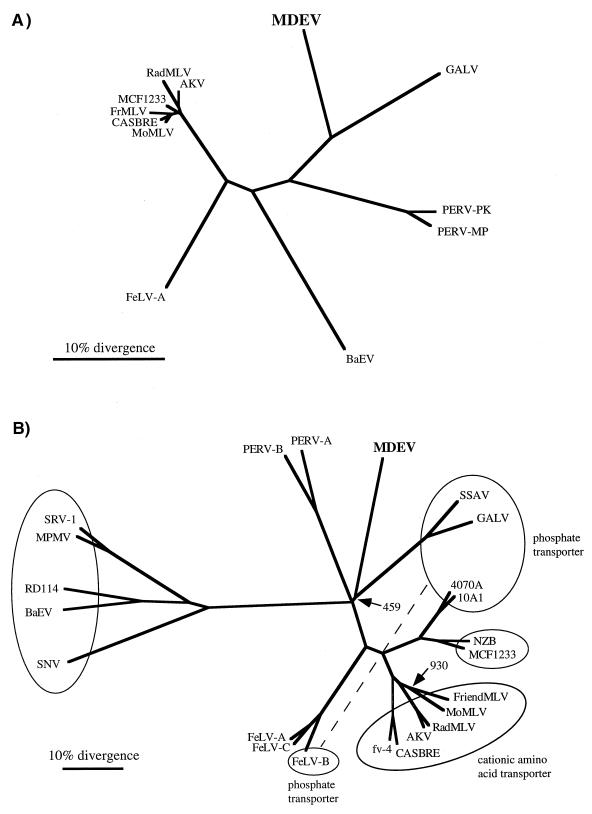
FIG. 4.
Phylogenetic trees based on Pol (A) and Env (B) proteins. The protein alignments, phylogenetic trees, and bootstrap analyses were performed with the Clustal W program, and the trees were viewed by using the TreeView program. (A) Tree based on PR-RT regions of Pol proteins. Only those positions of the alignment with no gaps were used to create the phylogenetic tree. All major branchpoints had confidence values of >98%, as measured by bootstrap analysis, except for branchpoints among the closely related members of the mouse virus group at the left. (B) Tree based on entire Env proteins. Viruses observed to share receptors in at least one cell type are grouped together in ovals, and the receptor is indicated in the cases where it is known. Note that feline leukemia virus type B (FeLV-B) belongs to the phosphate transporter group but cannot be included in that oval in a two-dimensional tree. MDEV and RD114 were not grouped due to the inconsistent nature of the interference observed in G355 cells. A phylogenetic tree created by excluding positions with a gap for any sequence was nearly identical. All major branchpoints had confidence values of >97%, as measured by bootstrap analysis, except for branchpoints indicated by the number of trials of the 1,000 that produced the displayed branchpoint. Abbreviations: AKV, AKR mouse ecotropic virus; BaEV, baboon endogenous virus; CASBRE, Casitas brain ecotropic virus; FrMLV, Friend MLV; NZB, New Zealand black xenotropic virus; RadMLV, radiation MLV; SNV, spleen necrosis virus; SRV-1, simian retrovirus type 1; SSAV, simian sarcoma-associated virus.