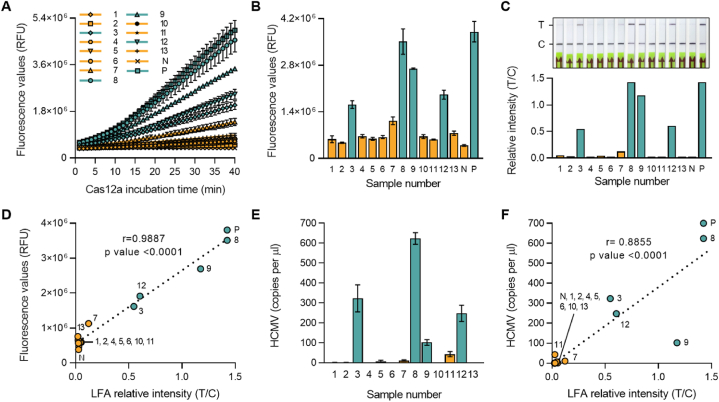
Fig. 6.
HCMV diagnosis of patient blood samples using RPA-CRISPR/Cas12a-LFA and qPCR. (A) Fluorescence signals were detected during the CRISPR-Cas12a reaction by a plate reader every minute. (B) Fluorescence values after 30 min of Cas12a reaction in panel A. Error bars indicate SD (n = 3). Comparisons between groups were performed using the two-tailed unpaired Student's t-test. (C) Diagnosis results were read out using an LFA strip after 30 min of CRISPR-Cas12a reaction. The photo was taken 3 min after the strips were dipped (top). Relative band intensity of the LFA strip (bottom). (D) Linear regression analysis between fluorescence values after 30 min of Cas12a reaction and relative band intensities of LFA strips. (E) Predicted viral copy numbers were calculated by qPCR. Error bars represent the results of three independent technical replicates. (F) Linear regression analysis between calculated viral copy numbers and relative band intensities of LFA strips. Green bars and dots indicate positive diagnosed patient samples, and yellow bars and dots indicate negative patient samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)