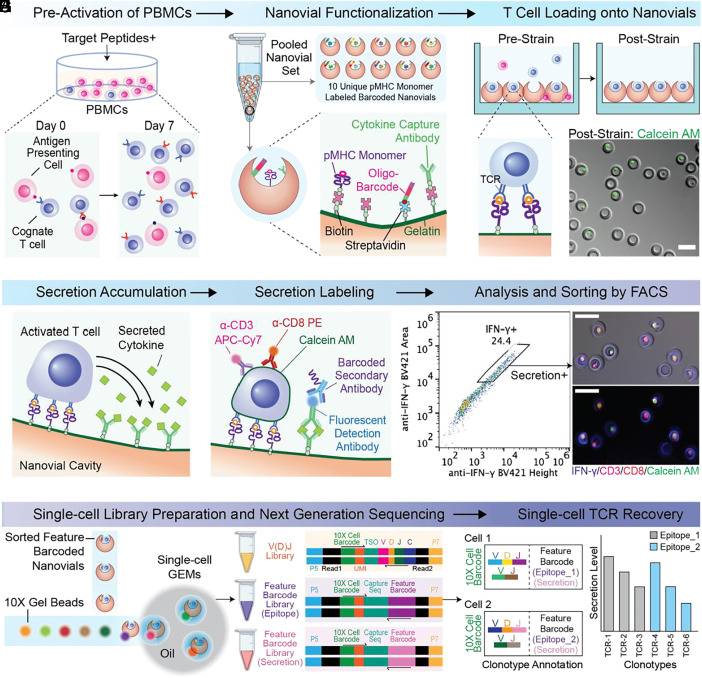
Fig. 1.
Overview of high-throughput analysis and isolation of antigen-specific T cells followed by recovery of a single-cell TCR library. (A) Optional pre-expansion of PBMCs with target peptides for 7 d. (B) Functionalization of nanovials with secretion capture antibodies, pMHC monomers, and oligonucleotide barcodes via streptavidin-biotin chemistry. (C) Loading of cognate T cells into the cavities of nanovials in a well plate and removal of unbound cells using a cell strainer. (D) Activation of T cells for 3 h and secretion capture in the cavity of nanovials. (E) Labeling of captured cytokines and cell surface markers with fluorescent detection antibodies, followed by oligonucleotide barcoded antibodies against secreted markers. (F) Sorting of cells on nanovials based on viability, CD3/CD8 expression, and secretion signal. (G) Compartmentalization of sorted population into droplets with a cell barcode bead in the 10X Chromium system for the construction of matched V(D)J and feature barcode libraries (nanovial-epitope and secretion barcodes). (H) Annotation of TCR clonotypes with corresponding secretion levels and epitopes by matching feature barcodes. (Scale bars represent 50 μm.)