Abstract
Genome-wide sequencing and genetic matchmaker services are propelling a new era of genotype-driven ascertainment of novel genetic conditions. The degree to which reported phenotype data in discovery-focused studies address informational priorities for clinicians and families is unclear. We identified reports published from 2017 to 2021 in 10 genetics journals of novel Mendelian disorders. We adjudicated the quality and detail of the phenotype data via 46 questions pertaining to six priority domains: (I) Development, cognition, and mental health; (II) Feeding and growth; (III) Medication use and treatment history; (IV) Pain, sleep, and quality of life; (V) Adulthood; and (VI) Epilepsy. For a subset of articles, all subsequent published follow-up case descriptions were identified and assessed in a similar manner. A modified Delphi approach was used to develop consensus reporting guidelines, with input from content experts across four countries. In total, 200 of 3243 screened publications met inclusion criteria. Relevant phenotypic details across each of the 6 domains were rated superficial or deficient in >87% of papers. For example, less than 10% of publications provided details regarding neuropsychiatric diagnoses and “behavioural issues”, or about the type/nature of feeding problems. Follow-up reports (n = 95) rarely contributed this additional phenotype data. In summary, phenotype information relevant to clinical management, genetic counselling, and the stated priorities of patients and families is lacking for many newly described genetic diseases. The PHELIX (PHEnotype LIsting fiX) reporting guideline checklists were developed to improve phenotype reporting in the genomic era.
Subject terms: Genetics research, Paediatric research
Introduction
Genome-wide sequencing and genetic matchmaker services have created a new paradigm for Mendelian disorder delineation1–3. Compared to prior decades, when syndrome identification was predominantly phenotype driven, there is now an increasing focus on “genomic ascertainment”4 [i.e. initially grouping individuals based on genomic variants of interest rather than (typically non-specific) phenotypes] and on generating functional evidence or usage of non-human model systems to support the disease-variant/gene association. Variability in the consistency of phenotyping and describing of findings is problematic. This variability can be exacerbated by individual sites each contributing only a single patient to an international case-series study, or the extraction of phenotype data from laboratory test requisitions. The field is converging around efforts to develop standardize terminology (e.g. Human Phenotype Ontology or HPO5, Medical Action Ontology6) and machine-readable, interoperable standards for recording and sharing phenotypes (e.g. Phenopacket Schema7–9). However, there are yet no well-defined nor broadly accepted minimum standards for phenotype descriptions of putative novel disorders with multisystem manifestations and/or a neurodevelopmental component.
After a first description of a novel Mendelian disorder is published, patients soon thereafter begin to be diagnosed via clinical genome-wide sequencing10–12. These individually ultra-rare13 conditions are collectively important contributors to the burden of genetic disease in the population14. The typical benefits of a molecular genetic diagnosis13,15 are attenuated when there is limited information available to inform genotype–phenotype correlation, natural history, prognostication, and anticipatory care. A key consideration in the assessment of ultra-rare conditions for potential “precision therapy” development is the degree to which the patient’s clinical trajectory can be anticipated16,17. Families who are among the first to receive a diagnosis of an ultra-rare genetic disorder have endorsed frustration with the perceived lack of information and support18,19. Similarly, clinicians face the same informational barrier, which impacts their abilities to care for and counsel patients and their families20.
Published expert opinions, survey data, reviews, and data from patient and family focus groups highlight key informational areas germane to the natural history of ultra-rare genetic diseases9,20–29. These informational areas have been studied in the context of more common genetic syndromes like Down syndrome, 22q11.2 deletion syndrome, fragile X syndrome, and the RASopathies30–38. We assessed the breadth and depth of phenotype reporting in contemporary descriptions of novel Mendelian genetic diseases across six priority domains: (I) Development, cognition, and mental health; (II) Feeding and growth; (III) Medication use and treatment history; (IV) Pain, sleep, and quality of life; (V) Adulthood; and (VI) Epilepsy. We also assessed in a similar manner follow-up reports appearing in the years following an initial report. These findings provided the impetus for, and guided the development of, the proposed new PHELIX (PHEnotype LIsting fiX) reporting guideline checklists, which complement other tools intended to improve phenotyping for rare genetic diseases5–9,39.
Results
Contemporary descriptions of new syndromes are often lacking in phenotype details
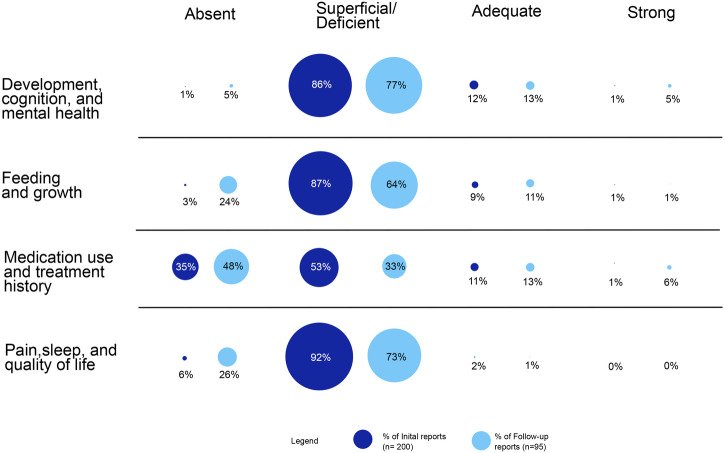
The 200 reports of 199 newly discovered genetic disorders included phenotype descriptions for a total of 1856 study participants (median: 7/report, range 2–42). Features of the reports (year and journal of publication) and of the participants (age) are summarized in Supplementary Tables 2 and 3). The overall qualitative assessment of reporting was deemed “superficial/deficient” or “absent” in 87% (Domain I: Development, cognition, and mental health) to 98% (Domain IV: Pain, sleep, and quality of life) of papers (Fig. 1). Five (2.5% of 200) reports were deemed “strong” in any single domain (pertaining to the genetic conditions associated with variants in the genes ADARB140, GNAI141, NCAPG242, PCDHGC443, and SPTBN144). No reports were deemed “strong” in their reporting across all Domains I–IV. The year and journal of publication were not associated with overall quality assessment of phenotype reporting (data not shown).
Fig. 1. Global qualitative assessments of the reporting of phenotype details germane to Domains I–IV.
For each of the four rating categories, the percentage of the initial reports (n = 200) are depicted in dark blue and of the follow-up reports (n = 95) in light blue. There were no significant differences in the distribution of overall quality ratings between the initial and follow-up reports, for any of the domains (Fisher’s exact tests, p > 0.05). See text for details.
Item-specific data supported the overall qualitative assessments of reporting quality (Supplementary Table 3 and Supplementary Figs. 3 and 4). While 97% of papers mentioned developmental concerns in study participants, 21% provided details about cognitive abilities for all the participants and a sole paper45 reported results from formal cognitive assessments for all participants (Supplementary Table 4). A common issue was that individuals were identified as having “developmental delay” without further elaboration. Similarly, of the papers that reported neuropsychiatric and behavioural issues in study participants, less than 5% of papers provided details for all participants regarding type/diagnosis, symptom severity, and/or nature of the assessments (Supplementary Table 3). Of the papers that reported on the presence of feeding difficulties, 8% consistently reported on the type/nature of feeding issues and current means of feeding (Supplementary Table 3). Growth parameters at birth were often reported, but 6% of papers reported on two or more growth measurements post-birth to allow for assessment of growth trajectories (Supplementary Table 4). Nearly half of all papers made no mention of participants’ medications or treatment trials, or of the absence thereof. The presence or absence of adverse effects of treatments were explicitly mentioned in just 21% of reports.
Domain V (Adulthood) was assessed in the subset of reports that included at least one adult individual (n = 63; adult defined as age ≥18 years) (Supplementary Fig. 1 and Supplementary Table 6). Domain VI (Epilepsy) was assessed in the subset of reports that included at least one study participant with seizures/epilepsy (n = 85) (Supplementary Fig. 1 and Supplementary Table 7). Consistent with the findings regarding Domains I–IV, most items were inconsistently or never reported (Supplementary Figs. 3 and 4). For example, papers rarely described proxies for adult functioning such as educational achievement or employment, nor the anti-seizure treatments for individuals with epilepsy.
Follow-up reports do not consistently address initial gaps in phenotype descriptions
Regarding the 25 genetic conditions first described in 2017, the 95 “follow-up” reports included phenotype descriptions for an additional 334 study participants (median: 1 per report, range 1–25). The overall qualitative assessment of reporting was similarly classified as “absent” or “superficial/deficient” in 81% (Domain III: Medication use and treatment history) to 99% (Domain IV: Pain, sleep, and quality of life) of papers (Fig. 1 and Supplementary Table 4), with no significant differences between the original and the follow-up reports. Eleven reports were deemed “strong” in any single domain (pertaining to the genetic conditions associated with variants in the eight genes CAMK2A46, CAMK2B46,47, DHX3048, OTUD6B49, PPP3CA50, UBTF51,52, WDR2653,54, and YY155). No reports were deemed “strong” in their reporting across each of Domains I–IV. Item-specific data are summarized in Supplementary Table 4.
Consensus phenotype reporting guidelines
Guideline checklists to enhance the reporting of phenotype data for ultra-rare genetic conditions were developed through a modified Delphi process56 and informed by the findings above (Supplementary Table 8). Specifically, items were included based on their superficial/deficient reporting in the literature to date, and on the recommendations of expert collaborators as being data that are both important to capture and feasible to obtain by researchers. The finalized checklist of 33 items across 9 categories is presented in Table 1 (PHELIX_General). To showcase how these guidelines could be expanded over time, additional items specific to epilepsy phenotype reporting are listed in Table 2 (PHELIX_Epilepsy). Extended versions with exemplar references are provided in Supplementary Tables 9 and 10. Examples of common deficiencies in phenotype reporting are provided in Supplementary Table 11.
Table 1.
Phenotype reporting checklist (PHELIX_General version 1.0)
| Phenotype category | Recommendation category | Specific recommendation for inclusion in report |
|---|---|---|
| 1. Development and cognition | Strongly recommended |
i. Standard and specific diagnostic term(s) for cognitive or developmental issue(s) ii. Level of cognitive functioning or degree of developmental delay iii. Age of attaining major milestones iv. Quantitative results from psychometric testing OR explicit acknowledgement that these results were not available |
| Optional, but encouraged | v. Narrative summary describing progression/change in cognitive or development issue(s) over time. | |
| 2. Behaviour and neuropsychiatric conditions | Strongly recommended | i. Standard and specific diagnostic term(s) for behavioural issues |
| Optional, but encouraged |
ii. Reported functional impact of behavioural/psychiatric condition iii. Age at diagnosis and/or age at first concern for behavioural issues iv. Impact of treatments/interventions, as reported by individuals, families, and/or clinicians |
|
| 3. Other medical conditions | Strongly recommended |
i. Major medical conditions ii. Presence or absence of issues in the following areas (if potentially associated with the condition under study): • Visual acuity and field of vision • Hearing ability • Speech/communication • Continence/toileting • Ambulation |
| 4. Feeding issues | Strongly recommended |
i. Functional impact of feeding issues ii. Current feeding method (e.g. oral, gastrostomy tube) |
| Optional, but encouraged |
iii. Age at first concern for feeding issues iv. Interventions and supports for feeding issues (e.g. feeding tube support) |
|
| 5. Growth | Strongly recommended |
i. Birth growth measurements AND gestational age-corrected centiles ii. Growth measurements (absolute values and z-scores) at two or more post-birth timepoints (where possible) |
| 6. Medication and treatment history | Optional, but encouraged |
i. Details of efficacious treatment(s) ii. Severe adverse events/reactions |
| 7. Pain, sleep, and quality of life | Optional, but encouraged |
i. Presence or absence of pain/neuroirritability ii. Presence or absence of abnormal sleep patterns/sleep disturbance iii. Qualitative description of proxies for quality of life, via patient and/or caregiver report iv. Direct assessment of quality of life using established measure(s), via patient and/or caregiver report |
| 8. Indicators of adult functional outcome | Strongly recommended |
i. Age at which the adult was last seen/phenotyped ii. Description of educational achievement iii. Nature of any employment (past and/or present) |
| Optional, but encouraged |
iv. Relationship status (past and/or present) v. Reproductive history |
|
| 9.Other | Strongly recommended |
i. Confirmation of informed consent to participate in the research study and to include the above phenotype information, for each participant ii. Distinguish between “not assessed” and “assessed and not present,” for every aspect of a phenotype described in the report and for each participant iii. Description of how phenotyping was performed (e.g. direct assessment by study team member(s), review of medical records, information provided on testing requisition), for each participant iv. Use of phenotype ontologies (e.g. HPO, ICD-11) and reporting tools (e.g. Phenopackets Schema) to standardize reporting, for each participant v. [For deceased participants] Cause of death |
HPO Human Phenotype Ontology, ICD-11 International Classification of Diseases 11th Revision.
Table 2.
Phenotype reporting recommendations for unprovoked seizures/epilepsy in individuals with newly described multisystem and/or neurodevelopmental Mendelian disorders (PHELIX_Epilepsy version 1.0)
| Phenotype subcategory | Recommendation category | Specific recommendation for inclusion in report |
|---|---|---|
| 1. Epilepsy syndrome and severity | Strongly recommended |
i. Seizure type(s) (per ILAE) ii.Age at seizure onset iii. EEG findings (including age(s) at time of study) iv. Epilepsy syndrome(s) (per ILAE) v. Findings that support the diagnosis of the epilepsy syndrome(s) (e.g. specific EEG findings) vi. Seizure frequency at last clinical assessment vii. Qualifiers of overall epilepsy severity (e.g. severe; treatment-refractory) |
| Optional, but encouraged |
viii. Clarification if EEG data were directly reviewed by members of the study team (versus only report details extracted from medical record) ix. Number of seizures requiring hospitalization in specific timeframe (e.g. last year) |
|
| 2. Pharmacological interventions | Strongly recommended |
i. Current and past medication name(s) ii. Perceived impact on seizure control |
| Optional, but encouraged |
iii. Dose iv. Duration of treatment trial v. Adverse effects/events due to the intervention |
|
| 3. Non-pharmacological interventions | Strongly recommended |
i. Intervention/procedure details (e.g. ketogenic diet, neurosurgery) ii. Perceived impact on seizure control |
| Optional, but encouraged | iii. Adverse effects/events due to the intervention | |
| 4. Brain imaging findings | Strongly recommended | i. Brain imaging findings (including age(s) at time of study) |
| Optional, but encouraged | ii. Clarification if brain imaging data were directly reviewed by members of the study team (versus only report details extracted from medical record) | |
| 5. Other | Optional, but encouraged |
i. Narrative summary of the progression of the individual’s seizure(s)/epilepsy phenotype over time ii. Narrative summary of the progression of the individual’s non-epilepsy phenotype over time (e.g. see PHELIX_General guidelines) |
EEG electroencephalogram, ILAE International League Against Epilepsy.
Discussion
Our results reveal that phenotype information relevant to clinical management, genetic counselling, and the stated priorities of patients and families, is lacking for many newly described genetic diseases. Although most published reports acknowledged the key phenotype domains assessed, few original or follow-up reports included clinically relevant details. To address this issue, we propose reporting guideline checklists for use by researchers and journals. Use of these guidelines could improve phenotype reporting in the era of genotype- and matchmaker service-driven reports of novel syndromes. Decision making about precision genetic or other therapy development, including the potential for N-of-1 trials, may be contingent on our understanding (or lack thereof) of the natural history of a given ultra-rare genetic disease16,17,57.
Reasons for under-reporting phenotype data are likely multiple and complex. First, these data may not be readily available to the referring clinician or laboratory collaborators, and “phenotyping is hard,”58 especially for older individuals with extensive past histories. Review of lifetime medical records, and/or a brief, targeted interview with patients and/or their caregiver(s), should be sufficient to gather most of the information outlined in Tables 1 and 2. Second, these data may not be requested by the coordinating research team that is leading the publication effort. In our experience, many groups design their own data collection forms that ask for no or only general details regarding issues outside of that group’s specific phenotype(s) of interest. Third, unlike for example DNA sequencing methods, there are no defined minimum reporting standards for phenotyping to guide peer reviewers and journal editors. Finally, there may be a belief that phenotype reporting in initial descriptions of novel genetic diseases is less important than establishing an association between variation in the gene and (any) disease phenotype. Although the hope may be that future reports will then describe many more individuals and include detailed phenotype data, we did not find evidence that this is consistently happening in practice in a timely manner.
We recognize several limitations of our review and guideline development methods. We selected only 10 top-tier genetics journals for our systematic review. The generalizability of our findings to reports published in other specialty-specific or organ system-specific journals is unclear. Out of necessity given the lack of validated tools, we created a new data collection questionnaire to assess the reporting of phenotype data and relied on subjective assessments from raters for some items. We selected broad phenotype domains based on our combined clinical experiences and the published literature; however, these domains are not the only important components of phenotyping. Ours was a paediatrics-focused effort, reflecting the phenotypes that are currently driving most Mendelian gene discovery efforts. Other groups may develop and add-on reporting criteria for additional specific phenotype elements, as we did for epilepsy, and continue to refine the general adult phenotype elements (Table 2). We also restricted our initial focus to cross-sectional reporting, and additional guidance will be needed for evaluating within-individual natural history. Finally, our reporting guidelines have not yet been applied prospectively to assess feasibility and utility.
We propose minimum standards for phenotype descriptions of putative novel disorders with multisystem manifestations and/or a neurodevelopmental component in children. Our intent is to encourage researchers to collect and share more details about key phenotypes where possible, recognizing that such efforts will be more challenging for some research groups and study designs (e.g. laboratory testing-based cohorts) than others. There are many forms of valid scholarship in the descriptions of rare diseases, and we strongly support multi-faceted approaches to phenotype delineation. Further refinement of our proposed reporting guidelines is an important consideration, including by collecting additional input from key stakeholders (e.g. rare disease organizations, journal editors). A key next step is to better integrate technologies for systematic phenotype collection and data sharing9,59. Our efforts were intended to be complementary to the Phenopackets Schema, and our findings provide further impetus for sharing phenotypic data in forms that are standardized and computable. Improved reporting of phenotype aspects like craniofacial morphology (dysmorphic features)60 and congenital anomalies could help with interpreting variants of uncertain significance and assessing phenotypic “fit”61,62. The aim of the PHELIX guideline checklists is to decrease the variability in the consistency of phenotyping and description of findings, and thereby enhance the ongoing clinical care of individuals with genetic conditions.
Methods
Systematic review
We utilized DistillerSR Version 2.35 for searching, screening, and data extraction (DistillerSR Inc, 2022; accessed January 2022–January 2023). We identified all first reports of novel genetic conditions discovered through genotype-driven ascertainment that result in multisystem and/or neurodevelopmental phenotypes, which were published in 1 of 10 genetics journals that are known for publishing novel reports of ultra-rare genetic conditions (American Journal of Human Genetics, American Journal of Medical Genetics Part A, Clinical Genetics, European Journal of Human Genetics, Genetics in Medicine, Genome Medicine, Human Molecular Genetics, Journal of Medical Genetics, Nature Genetics, PLoS Genetics) during a 5-year period (1 January 2017–31 December 2021). We selected this period because of the rise to prominence of “gene matchmaker” tools in the mid- to late-2010s, and to ensure feasibility of the systematic data extraction. The search executed on January 3, 2022, identified 3243 articles (Supplementary Table 1 and Supplementary Fig. 1). Exclusion criteria were: (i) prenatal or neonatal lethal phenotype, (ii) case report or description of a single family, (iii) new gene for a known clinical syndrome (e.g. Joubert syndrome, Noonan syndrome), (iv) chromosome disorder with non-recurrent breakpoints that did not definitively implicate a specific gene, (v) potential genotype–phenotype expansion rather than a novel disorder. We also excluded large-scale gene discovery efforts in populations with common complex diseases and/or clinical testing laboratory cohort studies, where the a priori expectation for detailed phenotype descriptions was low. After both title and abstract screening and full-text review stages, n = 200 reports describing 199 distinct monogenic conditions met the inclusion criteria (two reports of a novel condition were published at the same time; Supplementary Fig. 1). For the subset of 25 genetic conditions first described in 2017, we performed an additional search using DistillerSR on June 1, 2022 (Supplementary Table 1) to identify subsequent published case descriptions in any journal (total n = 95; Supplementary Fig. 2). Reference review and additional Internet searching did not identify any other “follow-up” reports.
Data extraction and analysis
For each published article (n = 295), study team members (authors A.A., A.J., A.P., M.Y.F.) adjudicated the phenotype data pertaining to six priority domains [(I) Development, cognition, and mental health; (II) Feeding and growth; (III) Medication use and treatment history; (IV) Pain, sleep, and quality of life; (V) Adulthood; and (VI) Epilepsy] using a custom designed data extraction form (Supplementary Tables 3, 5, and 6). Domains I–VI were included based on the study team’s clinical experience and review of the aforementioned published expert opinions, survey data, reviews, and data from patient and family focus groups9,20–29. Data for Domains V and VI were extracted separately from Domains I to IV. The total 46-item form was developed by members of the study team (authors A.A., A.J., C.D., N.J., D.B., G.C.) with clinical expertise in medical genetics, psychiatry, development, general paediatrics, paediatric palliative care, paediatric complex care, and paediatric hospitalist medicine. Each domain was associated with multiple issue-specific items. Descriptive statistics were calculated using Microsoft Excel.
A separate overall qualitative assessment of reporting quality (“strong”, “adequate”, “superficial/deficient”, “absent”, or “not applicable”) was also assigned for Domains I–IV. The overall qualitative assessment of reporting strength in each domain for each report was subjective and informed by data collected in the sub-questions (Supplementary Table 2). For the assessors, “absent” was defined as “no, or almost no, reporting of phenotype information [in this Domain]” and exemplified by reports with >80% of sub-questions having “No” or “Never” responses. “Superficial/deficient” was defined as “modest reporting of phenotype information [in this Domain], often lacking in detail and with concerning gaps from a genetic counselling perspective” and exemplified by reports with 50–80% of sub-questions having “No” or “Never” responses. “Adequate” was defined as “satisfactory reporting of phenotype information [in this Domain], in both breadth and depth, to facilitate genetic counselling and answer initial (common) clinician/family questions” and exemplified by reports with 20–50% of sub-questions having “No” or “Never” responses. “Strong” was defined as “intentional reporting of all available phenotype information [in this Domain], with a breadth and depth that would facilitate genetic counselling and answer initial and follow-up clinician/family questions” and exemplified by reports with <20% of sub-questions having “No” or “Never” responses. For reports with small numbers of participants, the percentage ranges did not apply. We confirmed high inter-rater reliability between the two independent assessors (>80% agreement) based on their blinded review of a subset of the same reports; discordant classifications were discussed together as a group to arrive at a consensus, before the raters proceeded to review the remainder of the reports independently.
Development of phenotype reporting guidelines through a modified Delphi process
Medical experts from member institutions of the International Precision Child Health Partnership (IPCHiP) participated in a modified Delphi process56. IPCHiP institutions included: Murdoch Children’s Research Institute/Royal Children’s Hospital (Melbourne, Australia), The Hospital for Sick Children (SickKids®; Toronto, ON, Canada), University College London/Greater Ormond Street Hospital (London, UK), and Boston Children’s Hospital (MA, USA)63,64. At the suggestion of the original study team members, additional expertise was sought in: (i) neuropsychological assessment and cognitive phenotyping (via Seaver Autism Center for Research and Treatment; NY, USA)65–67, and (ii) adult phenotyping (via University Health Network; Toronto, ON, Canada)68,69. Authors J.C., L.D., P.G., T.L., P.S., Z.S., J.A.S.V., C.D., N.J., and D.B. contributed to the initial refinement of guidelines for Domains I–V. We sent out three electronic surveys to the above authors (minimum engagement rate >50%) over a five-month period to define and prioritize the reporting criteria. We then hosted an online meeting that incorporated independent voting on inclusion/exclusion of each draft item. The meeting was recorded, to allow for asynchronous viewing by those expert volunteers who were unable to attend in real-time. Authors V.C., A.D., K.H., N.S.Y.L., A.T., A.P., and K.W. contributed to the initial refinement of guidelines for Domain VI (Epilepsy). Similarly, we used a series of two electronic surveys to define and prioritize the reporting criteria. All authors reviewed, revised, and ultimately approved the reporting guideline checklists for Domains I–VI reported herein. The guideline checklists will be uploaded to the EQUATOR (Enhancing the QUAlity and Transparency Of health Research) Network website (https://www.equator-network.org/) as the PHELIX_General (Table 1) and the PHELIX_Epilepsy checklists (Table 2).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Funding was provided by the SickKids Research Institute, the Canadian Institutes of Health Research (Funding Reference Number: PJT186240), and the University of Toronto McLaughlin Centre. IPCHiP is supported in part by SickKids Foundation donors including Jamie and Patsy Anderson and the Feiga Bresver Academic Foundation. D.B. also acknowledges the support of her research time at Holland Bloorview by the Arthur Family Foundation. J.A.S.V. holds the SickKids Psychiatry Associates Chair in Developmental Psychopathology at The Hospital for Sick Children. V.C. is supported by SickKids Foundation, Emma IS and the Margaret R. Anderson endowment. Research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. The Chair in Genomic Medicine awarded to J.C. is generously supported by The Royal Children’s Hospital Foundation. These funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author contributions
Conceptualization: D.B., G.C., methodology: A.A., A.J., C.D., N.J., D.B., G.C., software: N/A, validation: A.A., A.J., A.P., M.Y.F., G.C., formal analysis: A.A., investigation: A.A., A.J., A.P., M.Y.F., V.C., A.D., K.H., N.S.Y.L., A.T., A.P., K.W., A.S.B., J.C., L.D., P.G., T.L., P.S., Z.S., J.A.S.V., C.D., N.J., D.B., G.C., resources: G.C., data curation: N/A, writing—original draft: A.A., A.J., D.B., G.C., writing—review and editing: A.P., M.Y.F., V.C., A.D., K.H., N.S.Y.L., A.T., A.P., K.W., A.S.B., J.C., L.D., P.G., T.L., P.S., Z.S., J.A.S.V., C.D., N.J., visualization: A.A., A.J., supervision: L.D., P.G., G.C., project administration: A.A., A.J., D.B., G.C., funding acquisition: G.C.
Data availability
The datasets analysed during the current study are available from the corresponding authors on reasonable request.
Competing interests
D.B. has received research funds from MapLight Therapeutics. K.W. has consulted for Stoke Therapeutics. J.A.S.V. has served as a consultant for NoBias Therapeutics Inc. and has received speaker fees for Henry Stewart Talks Ltd. These relationships did not influence content of this manuscript but are disclosed for potential future considerations. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of members and their affiliations appears in the Supplementary Information.
Contributor Information
Danielle Baribeau, Email: dbaribeau@hollandbloorview.ca.
Gregory Costain, Email: gregory.costain@sickkids.ca.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-024-00408-w.
References
- 1.Bamshad MJ, Nickerson DA, Chong JX. Mendelian gene discovery: fast and furious with no end in sight. Am. J. Hum. Genet. 2019;105:448–455. doi: 10.1016/j.ajhg.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boycott KM, et al. A diagnosis for all rare genetic diseases: the horizon and the next frontiers. Cell. 2019;177:32–37. doi: 10.1016/j.cell.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 3.Boycott KM, Azzariti DR, Hamosh A, Rehm HL. Seven years since the launch of the Matchmaker Exchange: the evolution of genomic matchmaking. Hum. Mutat. 2022;43:659–667. doi: 10.1002/humu.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilczewski CM, et al. Genotype first: clinical genomics research through a reverse phenotyping approach. Am. J. Hum. Genet. 2023;110:3–12. doi: 10.1016/j.ajhg.2022.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargano, M. A. et al. The Human Phenotype Ontology in 2024: phenotypes around the world. Nucleic Acids Res. 10.1093/nar/gkad1005 (2023). [DOI] [PMC free article] [PubMed]
- 6.Carmody LC, et al. The Medical Action Ontology: a tool for annotating and analyzing treatments and clinical management of human disease. Med. 2023;4:913.e3–927.e3. doi: 10.1016/j.medj.2023.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danis D, et al. Phenopacket-tools: building and validating GA4GH Phenopackets. PLoS ONE. 2023;18:e0285433. doi: 10.1371/journal.pone.0285433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ladewig MS, et al. GA4GH Phenopackets: a practical introduction. Adv. Genet. 2023;4:2200016. doi: 10.1002/ggn2.202200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobsen JOB, et al. The GA4GH Phenopacket schema defines a computable representation of clinical data. Nat. Biotechnol. 2022;40:817–820. doi: 10.1038/s41587-022-01357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bick D, Jones M, Taylor SL, Taft RJ, Belmont J. Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases. J. Med. Genet. 2019;56:783–791. doi: 10.1136/jmedgenet-2019-106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Australian Genomics Health Alliance Acute Care Flagship et al. Feasibility of ultra-rapid exome sequencing in critically ill infants and children with suspected monogenic conditions in the Australian Public Health Care System. JAMA. 2020;323:2503–2511. doi: 10.1001/jama.2020.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costain G, Cohn RD, Scherer SW, Marshall CR. Genome sequencing as a diagnostic test. CMAJ. 2021;193:E1626–E1629. doi: 10.1503/cmaj.210549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costain G, et al. Genome sequencing as a diagnostic test in children with unexplained medical complexity. JAMA Netw. Open. 2020;3:e2018109. doi: 10.1001/jamanetworkopen.2020.18109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haque B, et al. Contemporary aetiologies of medical complexity in children: a cohort study. Arch. Dis. Child. 2023;108:147–149. doi: 10.1136/archdischild-2022-325094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costain G, Chow EW, Ray PN, Bassett AS. Caregiver and adult patient perspectives on the importance of a diagnosis of 22q11.2 deletion syndrome. J. Intellect. Disabil. Res. 2012;56:641–651. doi: 10.1111/j.1365-2788.2011.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pena LDM, et al. Contributions from medical geneticists in clinical trials of genetic therapies: a points to consider statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2023;25:100831. doi: 10.1016/j.gim.2023.100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleeson JG, et al. Personalized antisense oligonucleotides ‘for free, for life’ - the n-Lorem Foundation. Nat. Med. 2023;29:1302–1303. doi: 10.1038/s41591-023-02335-2. [DOI] [PubMed] [Google Scholar]
- 18.Fanos JH. New “first families”: the psychosocial impact of new genetic technologies. Genet. Med. 2012;14:189–190. doi: 10.1038/gim.2011.17. [DOI] [PubMed] [Google Scholar]
- 19.Lee W, et al. Genome sequencing among children with medical complexity: what constitutes value from parents’ perspective? J. Genet. Couns. 2022;31:523–533. doi: 10.1002/jgc4.1522. [DOI] [PubMed] [Google Scholar]
- 20.Vandeborne L, van Overbeeke E, Dooms M, De Beleyr B, Huys I. Information needs of physicians regarding the diagnosis of rare diseases: a questionnaire-based study in Belgium. Orphanet J. Rare Dis. 2019;14:99. doi: 10.1186/s13023-019-1075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diskin, C. et al. Research priorities for children with neurological impairment and medical complexity in high-income countries. Dev. Med. Child Neurol. 10.1111/dmcn.15037 (2021). [DOI] [PMC free article] [PubMed]
- 22.Nelson KE, et al. Clinical characteristics of children with severe neurologic impairment: a scoping review. J. Hosp. Med. 2023;18:65–77. doi: 10.1002/jhm.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill PJ, et al. Patient, caregiver, and clinician participation in prioritization of research questions in pediatric hospital medicine. JAMA Netw. Open. 2022;5:e229085. doi: 10.1001/jamanetworkopen.2022.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacquemont S, et al. Genes To Mental Health (G2MH): a framework to map the combined effects of rare and common variants on dimensions of cognition and psychopathology. Am. J. Psychiatry. 2022;179:189–203. doi: 10.1176/appi.ajp.2021.21040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Commission on Novel Technologies for Neurodevelopmental Copy Number Variants. Neurodevelopmental copy-number variants: a roadmap to improving outcomes by uniting patient advocates, researchers, and clinicians for collective impact. Am. J. Hum. Genet. 2022;109:1353–1365. doi: 10.1016/j.ajhg.2022.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanly C, Shah H, Au PYB, Murias K. Description of neurodevelopmental phenotypes associated with 10 genetic neurodevelopmental disorders: a scoping review. Clin. Genet. 2021;99:335–346. doi: 10.1111/cge.13882. [DOI] [PubMed] [Google Scholar]
- 27.McTague A, et al. Defining causal variants in rare epilepsies: an essential team effort between biomedical scientists, geneticists and epileptologists. Eur. J. Med. Genet. 2022;65:104531. doi: 10.1016/j.ejmg.2022.104531. [DOI] [PubMed] [Google Scholar]
- 28.Perlman P, et al. Support to caregivers who have received genetic information about neurodevelopmental and psychiatric vulnerability in their young children: a narrative review. Clin. Genet. 2023;104:163–173. doi: 10.1111/cge.14349. [DOI] [PubMed] [Google Scholar]
- 29.Baribeau DA, et al. Developmental implications of genetic testing for physical indications. Eur. J. Hum. Genet. 2022;30:1297–1300. doi: 10.1038/s41431-022-01181-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altmuller F, et al. Genotype and phenotype spectrum of NRAS germline variants. Eur. J. Hum. Genet. 2017;25:823–831. doi: 10.1038/ejhg.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armour CM, Allanson JE. Further delineation of cardio-facio-cutaneous syndrome: clinical features of 38 individuals with proven mutations. J. Med. Genet. 2008;45:249–254. doi: 10.1136/jmg.2007.054460. [DOI] [PubMed] [Google Scholar]
- 32.Bassett AS, et al. Clinical features of 78 adults with 22q11 deletion syndrome. Am. J. Med. Genet. A. 2005;138:307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fung WL, et al. Practical guidelines for managing adults with 22q11.2 deletion syndrome. Genet. Med. 2015;17:599–609. doi: 10.1038/gim.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapadia RK, Bassett AS. Recognizing a common genetic syndrome: 22q11.2 deletion syndrome. CMAJ. 2008;178:391–393. doi: 10.1503/cmaj.071300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kidd SA, et al. Fragile X syndrome: a review of associated medical problems. Pediatrics. 2014;134:995–1005. doi: 10.1542/peds.2013-4301. [DOI] [PubMed] [Google Scholar]
- 36.Korenberg JR, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl Acad. Sci. USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mautner VF, et al. Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J. Med. Genet. 2010;47:623–630. doi: 10.1136/jmg.2009.075937. [DOI] [PubMed] [Google Scholar]
- 38.Pasmant E, Vidaud M, Vidaud D, Wolkenstein P. Neurofibromatosis type 1: from genotype to phenotype. J. Med. Genet. 2012;49:483–489. doi: 10.1136/jmedgenet-2012-100978. [DOI] [PubMed] [Google Scholar]
- 39.Chung WK, et al. Genomic medicine implementation protocols in the PhenX Toolkit: tools for standardized data collection. Genet. Med. 2021;23:1783–1788. doi: 10.1038/s41436-021-01183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan TY, et al. Bi-allelic ADARB1 variants associated with microcephaly, intellectual disability, and seizures. Am. J. Hum. Genet. 2020;106:467–483. doi: 10.1016/j.ajhg.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muir AM, et al. Variants in GNAI1 cause a syndrome associated with variable features including developmental delay, seizures, and hypotonia. Genet. Med. 2021;23:881–887. doi: 10.1038/s41436-020-01076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan TN, et al. Mutations in NCAPG2 cause a severe neurodevelopmental syndrome that expands the phenotypic spectrum of condensinopathies. Am. J. Hum. Genet. 2019;104:94–111. doi: 10.1016/j.ajhg.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iqbal M, et al. Biallelic variants in PCDHGC4 cause a novel neurodevelopmental syndrome with progressive microcephaly, seizures, and joint anomalies. Genet. Med. 2021;23:2138–2149. doi: 10.1038/s41436-021-01260-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cousin MA, et al. Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nat. Genet. 2021;53:1006–1021. doi: 10.1038/s41588-021-00886-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harel T, et al. Homozygous stop-gain variant in LRRC32, encoding a TGFbeta receptor, associated with cleft palate, proliferative retinopathy, and developmental delay. Eur. J. Hum. Genet. 2019;27:1315–1319. doi: 10.1038/s41431-019-0380-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akita T, et al. De novo variants in CAMK2A and CAMK2B cause neurodevelopmental disorders. Ann. Clin. Transl. Neurol. 2018;5:280–296. doi: 10.1002/acn3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dwyer BK, Veenma DCM, Chang K, Schulman H, Van Woerden GM. Case report: Developmental delay and acute neuropsychiatric episodes associated with a de novo mutation in the CAMK2B gene (c.328G>A p.Glu110Lys) Front. Pharm. 2022;13:794008. doi: 10.3389/fphar.2022.794008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mannucci I, et al. Genotype-phenotype correlations and novel molecular insights into the DHX30-associated neurodevelopmental disorders. Genome Med. 2021;13:90. doi: 10.1186/s13073-021-00900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romero-Ibarguengoitia ME, et al. Comparison of genetic variants and manifestations of OTUD6B-related disorder: the first Mexican case. J. Investig. Med. High. Impact Case Rep. 2020;8:2324709620957777. doi: 10.1177/2324709620957777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizuguchi T, et al. Loss-of-function and gain-of-function mutations in PPP3CA cause two distinct disorders. Hum. Mol. Genet. 2018;27:1421–1433. doi: 10.1093/hmg/ddy052. [DOI] [PubMed] [Google Scholar]
- 51.Toro C, et al. A recurrent de novo missense mutation in UBTF causes developmental neuroregression. Hum. Mol. Genet. 2018;27:691–705. doi: 10.1093/hmg/ddx435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedlackova L, et al. UBTF mutation causes complex phenotype of neurodegeneration and severe epilepsy in childhood. Neuropediatrics. 2019;50:57–60. doi: 10.1055/s-0038-1676288. [DOI] [PubMed] [Google Scholar]
- 53.Cospain A, et al. Skraban-Deardorff syndrome: six new cases of WDR26-related disease and expansion of the clinical phenotype. Clin. Genet. 2021;99:732–739. doi: 10.1111/cge.13933. [DOI] [PubMed] [Google Scholar]
- 54.Pavinato L, et al. Expanding the clinical phenotype of the ultra-rare Skraban-Deardorff syndrome: two novel individuals with WDR26 loss-of-function variants and a literature review. Am. J. Med. Genet. A. 2021;185:1712–1720. doi: 10.1002/ajmg.a.62157. [DOI] [PubMed] [Google Scholar]
- 55.Carminho-Rodrigues MT, et al. Complex movement disorder in a patient with heterozygous YY1 mutation (Gabriele-de Vries syndrome) Am. J. Med. Genet. A. 2020;182:2129–2132. doi: 10.1002/ajmg.a.61731. [DOI] [PubMed] [Google Scholar]
- 56.Moher D, Schulz KF, Simera I, Altman DG. Guidance for developers of health research reporting guidelines. PLoS Med. 2010;7:e1000217. doi: 10.1371/journal.pmed.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juando-Prats C, et al. DRAVET ENGAGE. Parent caregivers of children with Dravet syndrome: perspectives, needs, and opportunities for clinical research. Epilepsy Behav. 2021;122:108198. doi: 10.1016/j.yebeh.2021.108198. [DOI] [PubMed] [Google Scholar]
- 58.Collins C. Phenotype with a side of genotype, please: Patients, parents and priorities in rare genetic disease. Appl. Transl. Genom. 2016;8:42–44. doi: 10.1016/j.atg.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girdea M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum. Mutat. 2013;34:1057–1065. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 60.Biesecker LG. An introduction to standardized clinical nomenclature for dysmorphic features: the Elements of Morphology project. BMC Med. 2010;8:56. doi: 10.1186/1741-7015-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elliott AM, et al. Genome-wide sequencing and the clinical diagnosis of genetic disease: the CAUSES study. HGG Adv. 2022;3:100108. doi: 10.1016/j.xhgg.2022.100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mak CC, et al. Exome sequencing for paediatric-onset diseases: impact of the extensive involvement of medical geneticists in the diagnostic odyssey. NPJ Genom. Med. 2018;3:19. doi: 10.1038/s41525-018-0056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morton SU, et al. Multicenter consensus approach to evaluation of neonatal hypotonia in the genomic era: a review. JAMA Neurol. 2022;79:405–413. doi: 10.1001/jamaneurol.2022.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Gama AM, et al. Evaluation of the feasibility, diagnostic yield, and clinical utility of rapid genome sequencing in infantile epilepsy (Gene-STEPS): an international, multicentre, pilot cohort study. Lancet Neurol. 2023;22:812–825. doi: 10.1016/S1474-4422(23)00246-6. [DOI] [PubMed] [Google Scholar]
- 65.Levy T, et al. CHAMP1 disorder is associated with a complex neurobehavioral phenotype including autism, ADHD, repetitive behaviors and sensory symptoms. Hum. Mol. Genet. 2022;31:2582–2594. doi: 10.1093/hmg/ddac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levy T, et al. Strong evidence for genotype-phenotype correlations in Phelan-McDermid syndrome: results from the developmental synaptopathies consortium. Hum. Mol. Genet. 2022;31:625–637. doi: 10.1093/hmg/ddab280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang L, et al. Prospective and detailed behavioral phenotyping in DDX3X syndrome. Mol. Autism. 2021;12:36. doi: 10.1186/s13229-021-00431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butcher NJ, et al. Functional outcomes of adults with 22q11.2 deletion syndrome. Genet. Med. 2012;14:836–843. doi: 10.1038/gim.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malecki SL, et al. A genetic model for multimorbidity in young adults. Genet. Med. 2020;22:132–141. doi: 10.1038/s41436-019-0603-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analysed during the current study are available from the corresponding authors on reasonable request.