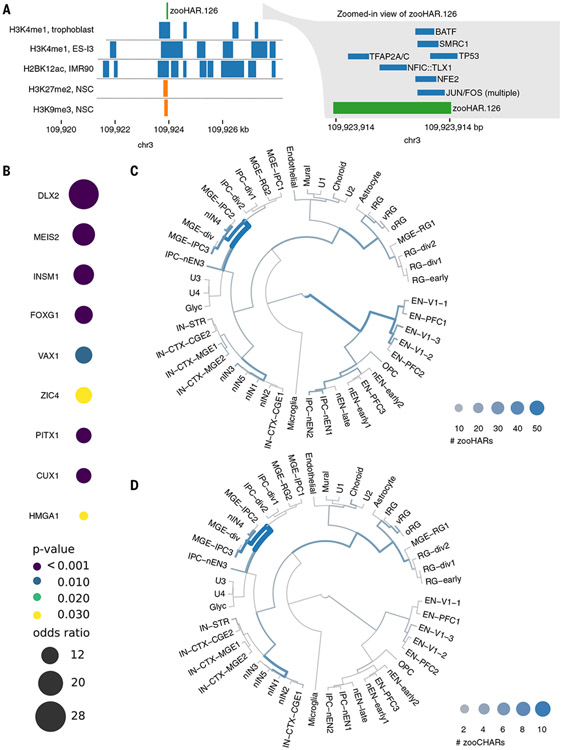
Fig. 3. zooHARs in human brain development.
(A) Transcription factor footprints (55) and epigenomic marks (59) overlapping zooHAR.126. NSC, neural stem cell. (B) Subset of enriched transcription factor footprints in zooHARs relative to phastCons elements (Fisher’s exact P ≤ 0.05). Full set is available in table S8. (C) Cell types in which zooHARs are predicted to regulate gene expression based on CellWalker analysis of data from the developing human telencephalon. (D) Cell type assignments for zooCHARs based on CellWalker analysis of data from the developing human telencephalon. Unlike with HARs, no CHARs map to late-stage excitatory neurons. Cell types are abbreviated as follows: excitatory neurons (ENs) derived from primary visual cortex (V1) or prefrontal cortex (PFC), newborn excitatory neurons (nENs), inhibitory cortical interneurons (IN-CTXs) originating in the medial/caudal ganglionic eminence (MGE/CGE), newborn interneurons (nINs), intermediate progenitor cells (IPCs), and truncated/ventral/outer radial glia (tRG/vRG/oRG). More cell type information is available at hhttps://cells.ucsc.edu/?ds=cortex-dev (58, 66).