Figure 3. Illustration of different redox-dependent chemistries involved in RNA modifications.
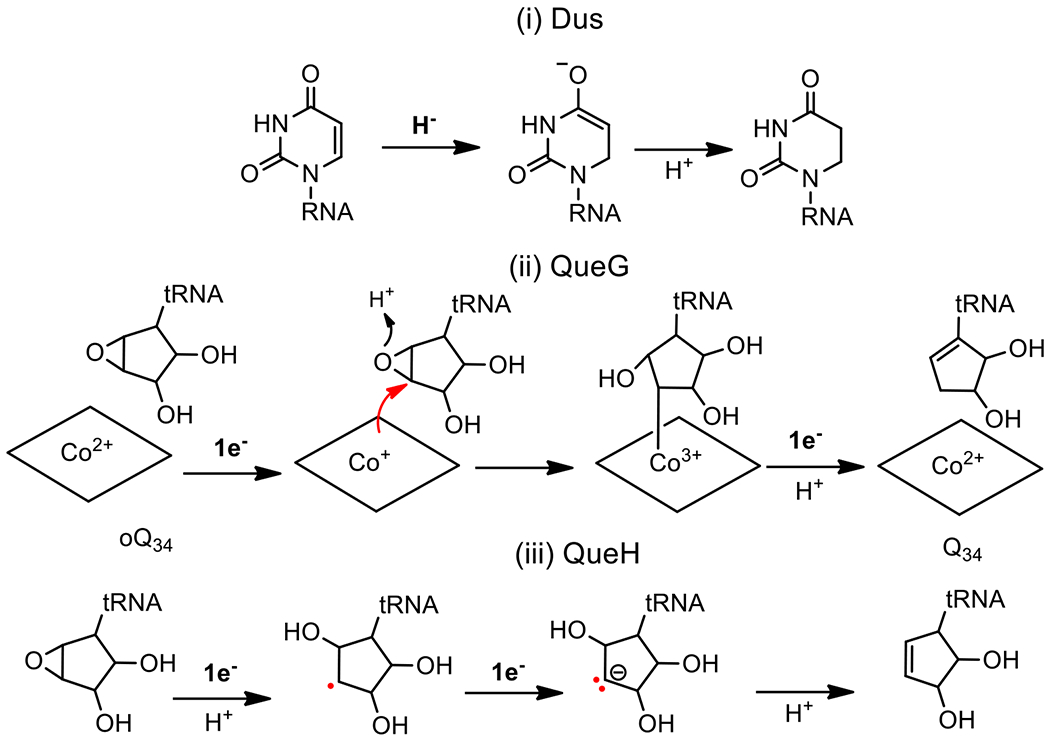
(i) Hydride reduction: Dus catalyzes the reduction of the ethylenic uridine bond of tRNAs and mRNAs by hydride transfer derived from the N5-FMNH− to the sp2 C6 carbon of uracil and by protonating the C5 using the thiol of a conserved site cysteine that acts as an acid. (ii) Covalently catalyzed reduction: epoxyqueuosine reductase, QueG, is the predominant oQ34 enzyme in nature that catalyzes the reduction of the epoxy-cyclopentanediol moiety of oQ34 to the cyclopentenediol of Q34 by covalent cobalamin-dependent catalysis. The reduction requires two x 1e- from the 4Fe-4S clusters of the protein while an aspartic acid from the active site functions as an acid. (iii) Reduction by electron transfer: QueH is the alternative enzyme to QueG but proceeds via a different mechanism based on sequential electron transfer from the single 4Fe-4S center of the protein.
