Figure 4. Strategy for the activation of an apoprotein version of a folate and flavin-dependent methyltransferase by a synthetic biomimetic for reductive methylation of tRNA.
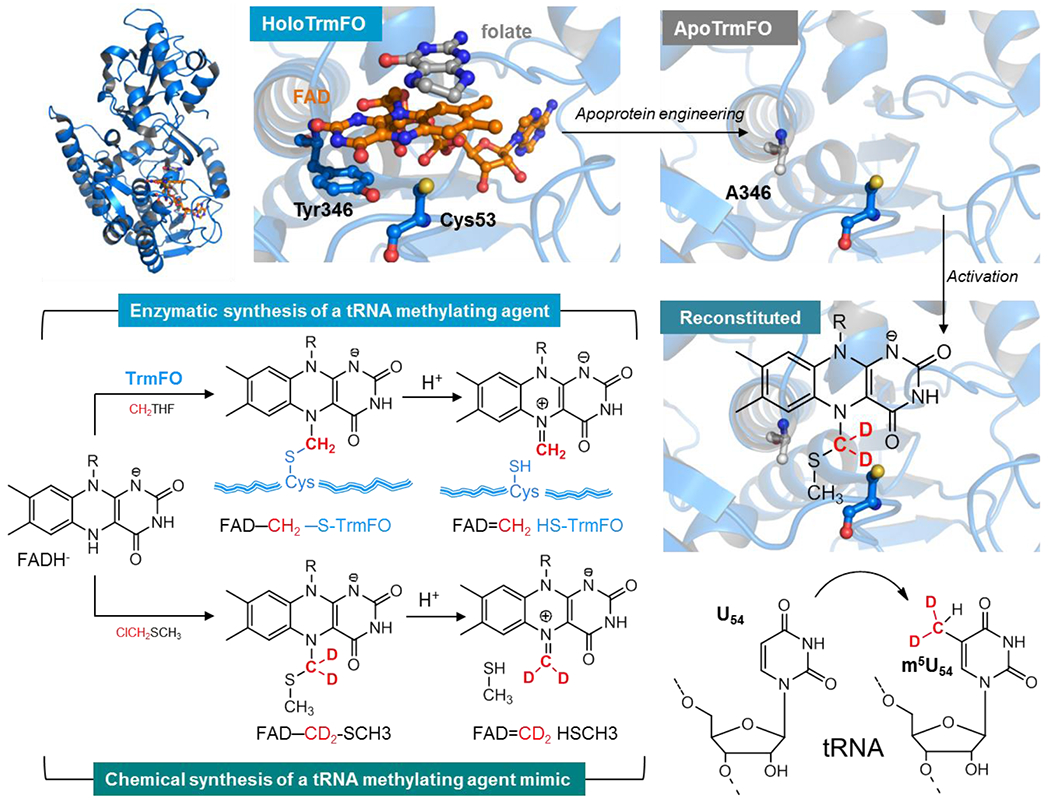
Top right shows the crystal structure of T. thermophilus TrmFO in complex with tetrahydrofolate (gray) (PDB: 3G5R). The zoom on a section of the active site shows the FAD coenzyme in orange, the folate derivative (in grey) whose pteridine stacks with the si-face of the FAD isoalloxazine while on the re-side lies the conserved tyrosine Y346 (numbering based on the sequence of BsTrmFO) engaged in π−π interaction with the flavin. The active site cysteine that stabilizes the flavin methylating intermediate FAD-CH2-S-CH3 (copurified with the Bs enzyme) faces the N5-FAD. This intermediate decomposes under protonation of the sulfur atom to give the FAD=CH2 species the bona fide C5-U methylating agent. Top left is a model of the flavin site of TtTrmFO showing the Y346A mutation artificially generated in PyMOL to illustrate the apoprotein. This apoprotein is reconstituted with the synthetic biomimetic flavin that acts as methylating agent and with the methylene deuterated to track its transfer on the substrate. Once reconstituted, the apoenzyme is activated for specific reductive methylation of the C5-U54 tRNA. This is shown by the incorporation of CD2 in red into the tRNA and its conversion to a methyl group via hydride transfer from the flavin to CHD2.
