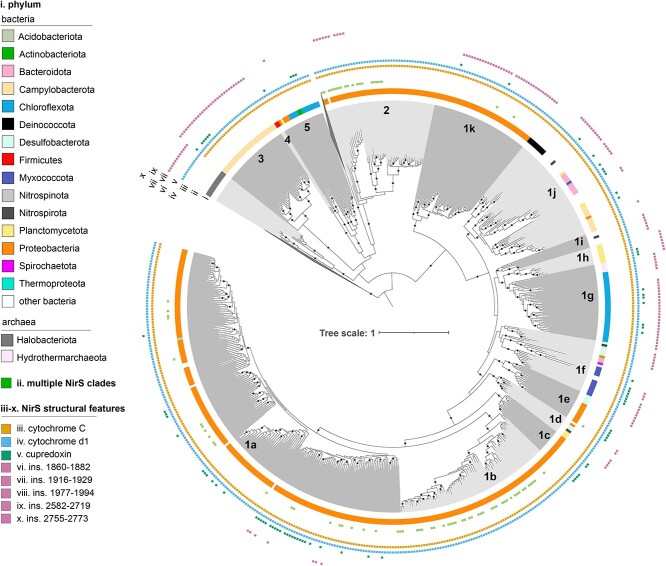
Figure 2.
Maximum-likelihood NirS phylogeny. The phylogeny was inferred from 542 full-length NirS amino acid sequences using the LG+F+R9 model in IQ-TREE. Black circles on tree indicate nodes with SH-aLRT ≥ 80% and Ufboot ≥ 95% support. Tips of tree are shaded according to clade, and data rings denote (i) phylum determined using GTDB-TK; (ii) presence of secondary nirS copies from other clades; (iii–x) select NirS structural features. The two outgroups, consisting of NirN and NirF sequences, are collapsed for clarity. Clades are numbered and lettered following Wei et al. [29] and Bonilla-Rosso et al. [28] to the degree possible, starting with the “canonical” Clade 1a [48].