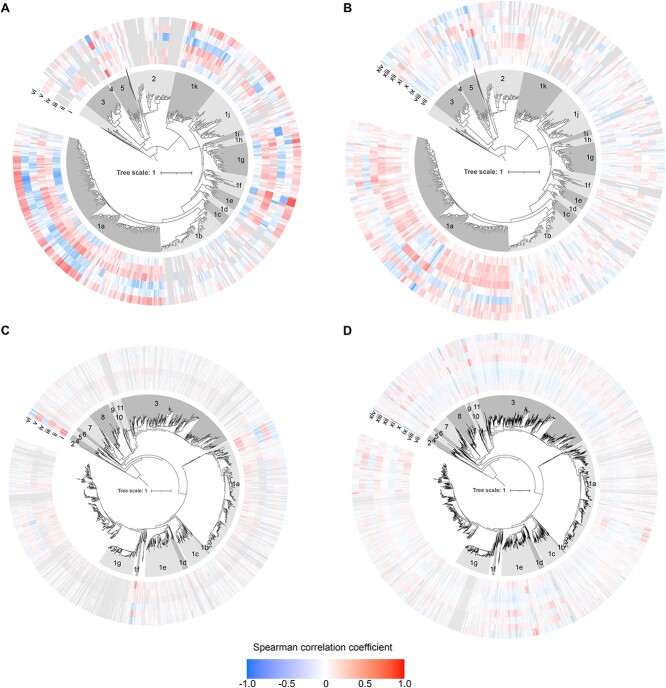
Figure 5.
Environmental correlations of nitrite reductases and their clades. Spearman correlation of edge masses on A, B) NirS and C, D NirK phylogenies against environmental variables in A, C) water column seawater and B, D) terrestrial biomes. Values represent correlations in leaves of tree. Outer rings are coloured according to correlation strength, with grey indicating insufficient data. (i) water nitrate + nitrite, (ii) water ammonium, (iii) chlorophyll, (iv) dissolved oxygen, (v) water depth, (vi) water temperature, (vii) soil nitrate, (viii) soil ammonium, (ix) percent SOC, (x) soil moisture, (xi) pH in CaCl2, (xii) sand content, (xiii) copper, and (xiv) iron. These variables were selected for having data available from many metagenomes and were potentially relevant for structuring microbial communities (reactive N availability (i, ii, vii, viii), C availability or input (iii, ix), oxygenation degree (iii, iv, x), and physiochemical properties (v-vi, xi-xiv). Bootstrap support values have been removed for visual clarity. Metagenomes with >15 placements for the gene were included. Since not all metadata are available for all samples, correlations correspond to different subsets of samples [48].