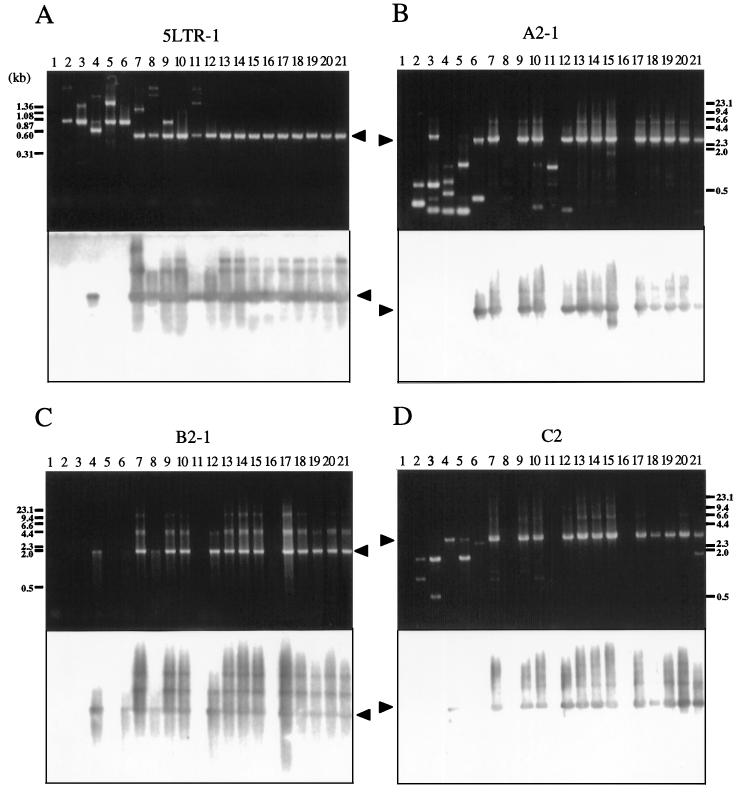
FIG. 4.
Analysis of BLV proviral genomes in BLV-infected cattle by conventional PCR. Each panel shows an ethidium bromide-stained gel (above) and patterns of hybridization (below) of PCR products in regions 5LTR-1 (A), A2-1 (B), B2-1 (C), and C2 (D). Genomic DNA from healthy cattle (lanes 2 to 12) and from cattle with EBL (lanes 13 to 21) was amplified by nested or seminested PCR with the primers shown in Fig. 2. Amplification was achieved by 30 or 35 cycles at 94°C for 30 s, 62°C for 20 s, and 72°C for 1 to 5 min, followed by a 5-min extension at 72°C. A second or third PCR was performed for 30 cycles with 1-μl samples from the first or second amplification. PCR products were separated on 2.0% (A) or 0.8% (B to D) agarose gels and hybridized with a full-length BLV probe as described in the legend to Fig. 1. Products derived from BLV proviral genomes are indicated by arrowheads. The positions of DNA molecular size markers (HaeIII-digested φX174 DNA [A] and HindIII-digested λ DNA [B to D]) are indicated. Lanes 1, BLV-negative animal 276; lanes 2, A30; lanes 3, A37; lanes 4, A42; lanes 5, A43; lanes 6, A50; lanes 7, A51; lanes 8, A52; lanes 9, A60; lanes 10, A78; lanes 11, S1; lanes 12, K67; lanes 13, pr1698; lanes 14, pr1720; lanes 15, pr2108; lanes 16, pr2119; lanes 17, pr2169; lanes 18, pr2374; lanes 19, pr2436; lanes 20, pr2501; lanes 21, pr2506.