Fig. 3. Chromatin accessibility and gene expression in 36-hour pupa forewing sections.
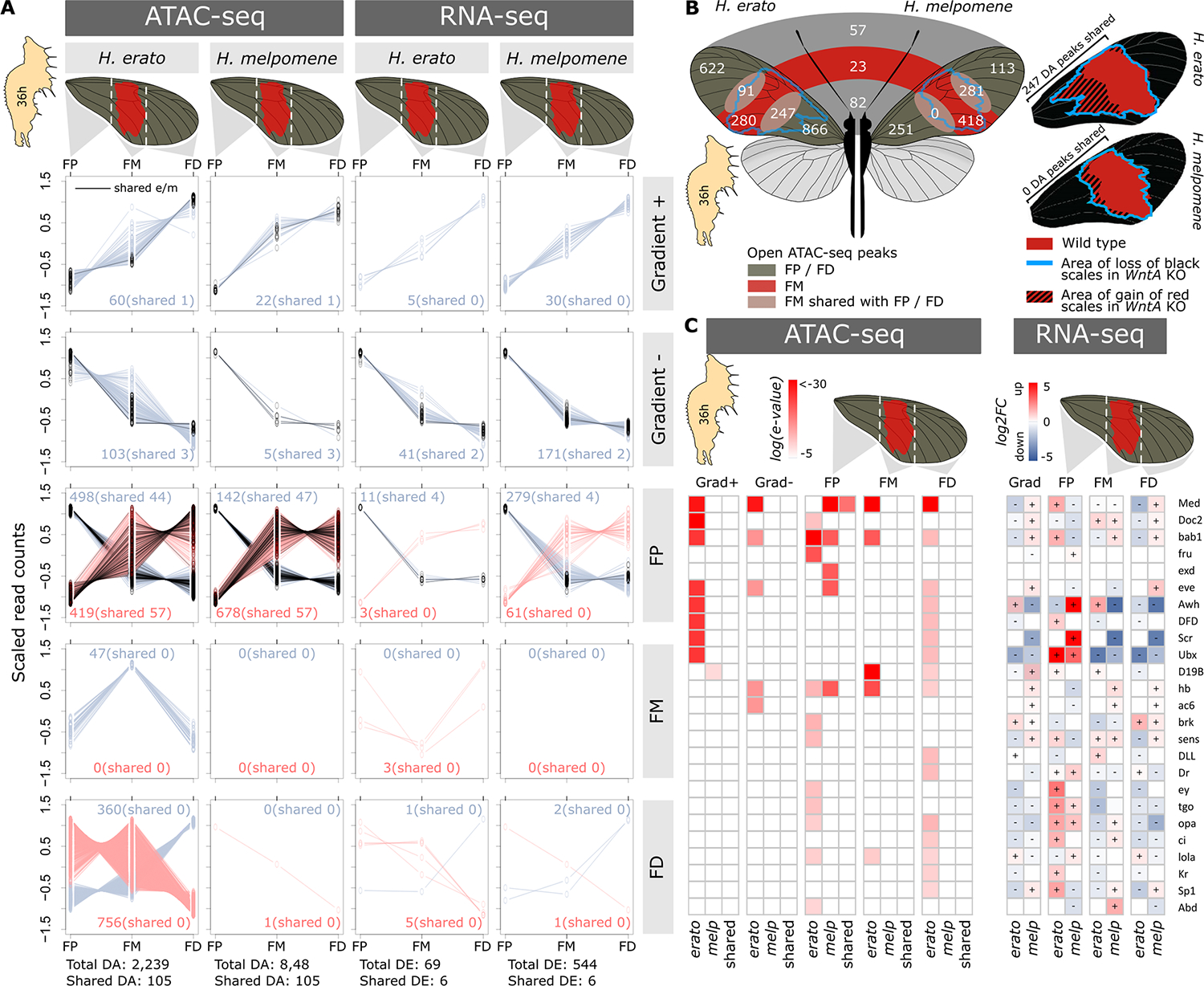
(A) Differentially accessible (DA) ATAC-seq peaks between forewing sections in H. erato and H. melpomene. ATAC-seq peaks are either significantly open (black lines) or closed (dark red lines) in FP, FM, FD, or a gradient + to − (increasing or decreasing accessibility from the proximal to distal wing section). Green lines indicate ATAC-seq peaks that are considered shared between H. erato and H. melpomene. For each comparison, we present the total and shared count numbers. (B) Numbers are differentially accessible ATAC-seq peaks in the wing sections. In contrast to (A), these numbers are obtained by pairwise comparisons between wing sections. Numbers at the boundaries of wing sections indicate peaks with shared differential accessibility compared to the other wing section. Numbers in the middle of the wings indicate peaks identified as shared between H. erato and H. melpomene (50% reciprocal overlap). Wings on the right show the wild-type phenotypes of H. erato and H. melpomene, with the blue lines indicating the extent of red scale development (and optix expression) in the WntA CRISPR-Cas9 KO. Numbers next to the wings represent DA peaks between FP or FM and FD in H. erato and H. melpomene, respectively. (C) TF motif enrichment (left) for differentially accessible ATAC-peaks between wing sections and expression of associated TFs (right). Log(e-value) indicates the significance level of the enrichment signal, and log2FC indicates the expression level relative to all other sections.
