Figure 6 .
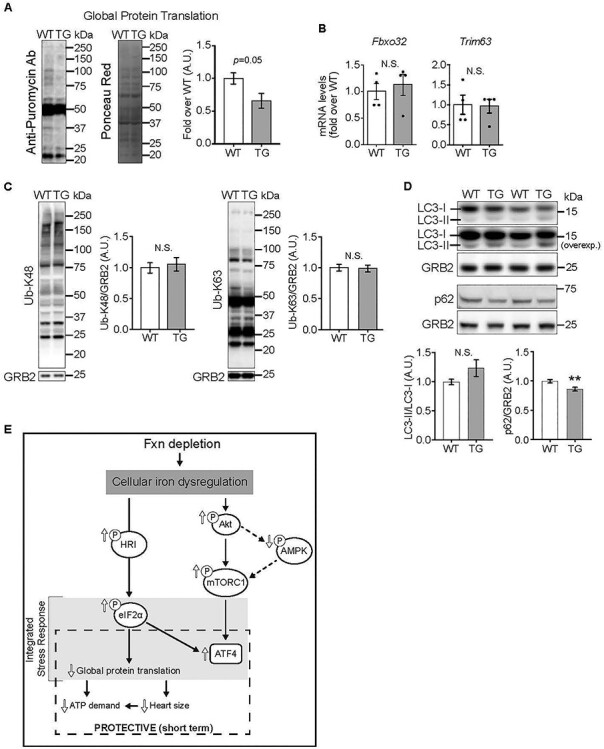
Decreased global protein translation in hearts depleted of FXN for 18 weeks (A) Global protein translation was determined using the SUnSET method. Left: Representative immunoblot using anti-puromycin antibody, with Ponceau Red staining (shown in gray scale) to estimate protein loading. Right: Quantification of puromycin signal (P = 0.05 by unpaired t-test, n = 4/genotype). (B, C) Protein degradation, estimated by relative mRNA levels of Fbxo32 and Trim63 (B, n = 4/genotype, not significant (N.S.), by unpaired t-test) and by K48- and K63-linked ubiquitination of proteins by immunoblot (C; N.S. by unpaired t-test, n = 6/genotype). (D) Autophagy, estimated by levels of LC3 and lipidated LC3 (n = 4/genotype, N.S.) and levels of p62 (n = 4/genotype, **P < 0.01), all by immunoblot. Statistical comparison was by unpaired t-test. (E) Model of altered signaling in FXN-depleted hearts. All values are the mean ± SEM.
