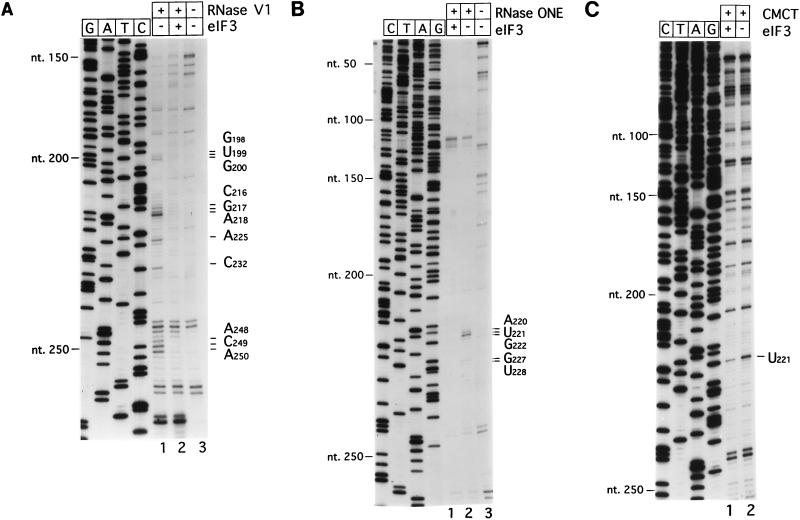
FIG. 2.
Chemical and enzymatic footprinting of the eIF3-CSFV IRES complex. Polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension shows the sensitivity of CSFV RNA upstream of nt 265 to cleavage by RNase V1 (lanes 1 and 2) either alone (lane 1) or complexed with eIF3 (lane 2) (A), the sensitivity of CSFV RNA upstream of nt 259 to cleavage by RNase ONE (lanes 1 and 2) either alone (lane 2) or complexed with eIF3 (lane 1) (B), and the reactivity of CSFV RNA upstream of nt 252 to modification by CMCT (lanes 1 and 2) either alone (lane 2) or complexed with eIF3 (lane 1) (C). cDNA products obtained after primer extension of untreated CSFV RNA are shown in lanes 3 of panels A and B. A dideoxynucleotide sequence generated with the same primer was run in parallel on each gel. The positions of protected residues are indicated to the right of each panel, and the positions of CSFV nucleotides at 50-nt intervals are indicated to the left of each panel.