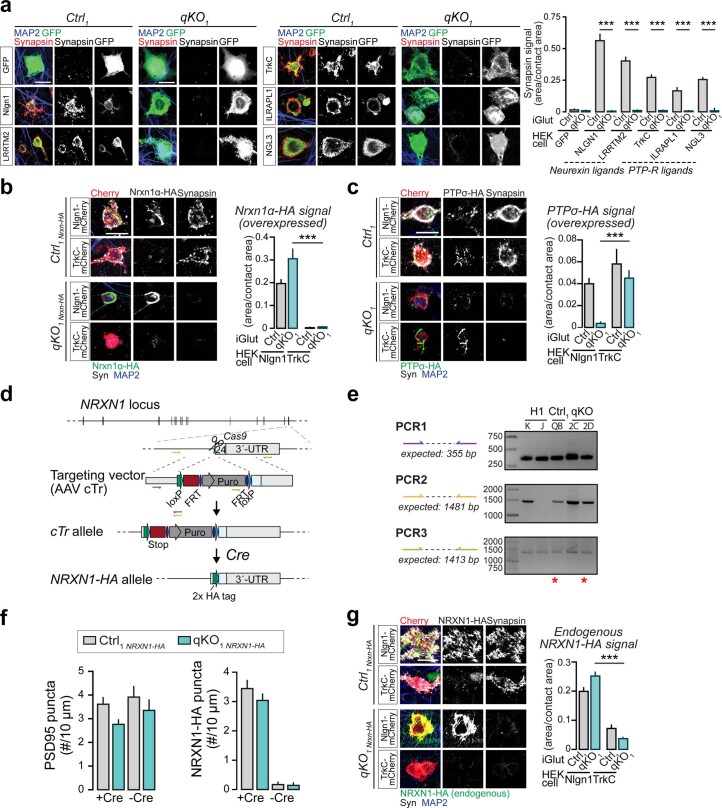
Extended Data Fig. 3. Analysis of presynaptic CAM pathways by heterologous synapses, insertion of an HA-tag in NRXN1 by homologous recombination and analysis of endogenous NRXN1 distribution (related to Fig. 2).
(a) Analysis of heterologous synapse formation by different pathways. Left. Representative images of HEK293 cells expressing the indicated GFP-tagged CAMs and co-cultured with control (Ctrl) or knockout (qKO) iGluts. Right. Quantitative analysis of synapsin recruitment to HEK293 cell areas. Number of cells/batches: 44-130/1 (see Supplementary Table 1). All conditions were repeated at least once with similar results. (b) Recruitment of overexpressed Nrxn1α-HA onto Nlgn1- or TrkC-expressing HEK293 cells. Left. Representative images. Right. Quantitative analysis of HA-recruitment to HEK293 cell areas. Number of cells/batches: 23-164/2-4 (see Supplementary Table 1). (c) Recruitment of overexpressed HA-tagged PTPσ onto Nlgn1- or TrkC-expressing HEK293 cells. Left. Representative images. Right. Quantitative analysis of Ctrl and qKO HA signal area onto HEK293 cells. Intracellular CASK–liprin-α interactions74 may explain the partial recruitment of PTPσ-HA by Nlgn1 in control cells. Number of cells/batches: 80-161/3-4 (see Supplementary Table 1). (d) Schematic representation of the targeting strategy to insert an HA-tag in the last exon of the NRXN1 gene. Homologous recombination yields a conditional truncation allele (cTr) producing soluble and rapidly degraded53 HA-tagged Neurexin-1. Cre- recombination restores expression of the wildtype, HA-tagged protein. (e) PCRs to confirm correct NRXN1 targeting in control and knockout clones, respectively. Color-coded primer pairs are shown in (d). Wildtype H1 cells were included as positive control for gene editing. Clones selected for further experiments marked by asterisks. (f) Quantification of PSD95 (left) and NRXN1-HA puncta densities (right) in Ctrl and qKO clones knock-in for NRXN1-HA (related to Fig. 2f). Number of dendrites/batches: 86-281/2 (see Supplementary Table 1). (g) Recruitment of endogenous HA-tagged NRXN1 onto Nlgn1- or TrkC-expressing HEK293 cells. Left. Representative images. Right. Quantitative analysis of Ctrl and qKO HA signal area onto HEK293 cells. Number of cells/batches: 90-239/2-3 (see Supplementary Table S1). Data in summary plots is represented as means ± SEM; *** p < 0.001.