Fig. 6. Sleep is required for mitophagy in neurons and glia.
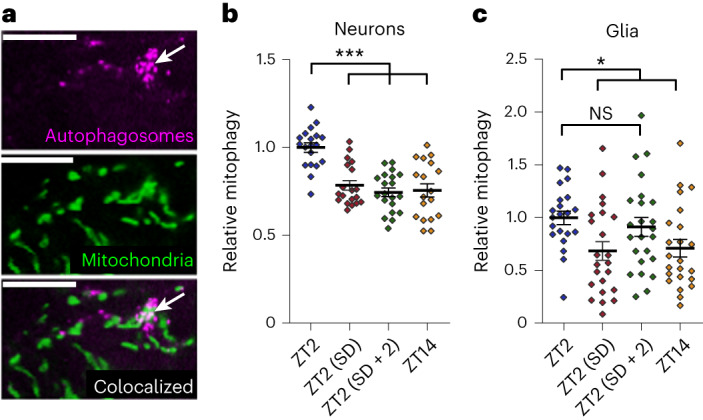
a, The mitochondrial and autophagy markers UAS-mito-GFP (middle) and UAS-Atg8-mCherry (top), respectively, were expressed in neurons with nSyb-GAL4 or glia with repo-GAL4. Colocalization (bottom) between autophagosomes and mitochondria was used to quantify mitophagy. The example images shown are from a small section of a single z slice of a brain with glial marker expression. The arrow indicates colocalization between mitochondria and autophagosomes (mitophagy); scale bars, 10 μm. b,c, Neuronal (b) and glial (c) mitophagy are greatest in the morning (ZT2) following a full night of sleep. Reduced mitophagy following a day of wake (ZT14) or a night of sleep deprivation (ZT2 (SD)) indicates that changes in mitophagy are sleep dependent rather than circadian clock dependent. Sleep deprivation/recovery and circadian time of fly collection are the same as shown in Fig. 1. For all data shown, *P < 0.05, **P < 0.01 and ***P < 0.001, while some groups with P > 0.05 (not significant) are unmarked. Bars/error bars indicate mean and s.e.m., respectively. Data points indicate individual brains/flies. The following are the numbers of flies (n) as plotted from left to right and statistical tests used: n = 19, 19, 20 and 18, Kruskall–Wallis test with a Dunn’s multiple testing correction (b); n = 21, 23, 24 and 23, Kruskall–Wallis test with a Dunn’s multiple testing correction (c).
