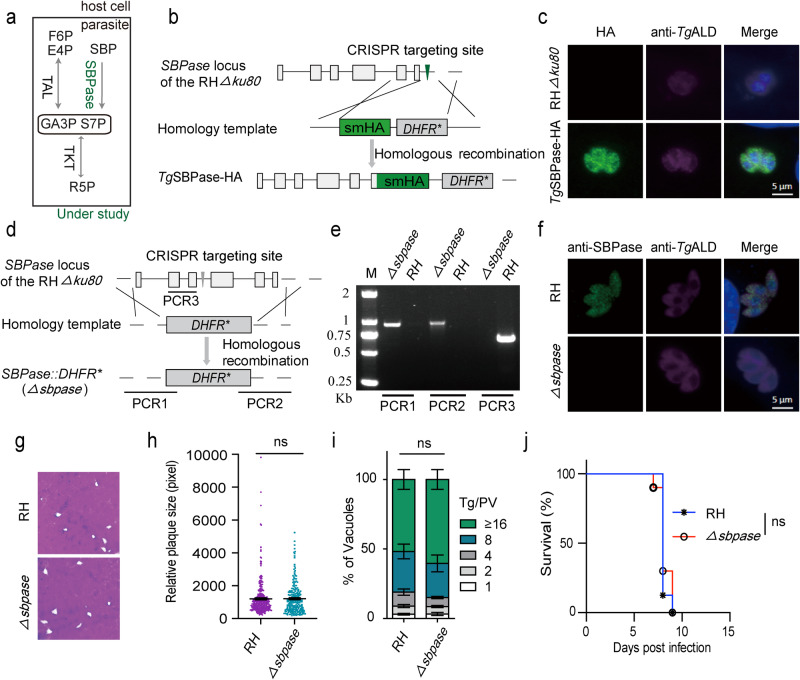
Fig. 1. SBPase is dispensable for parasite growth.
a Scheme of the R5P synthesis pathway in T. gondii, highlighting the SBPase enzyme. b Illustration depicting the 3′-genomic tagging of SBPase with a smHA epitope. c Co-localization of TgSBPase-HA and TgALD in tachyzoites. The C-terminal smHA tagging of SBPase was achieved by CRISPR/Cas9-assisted site-specific integration in the RHΔku80 strain. Scale bars = 5 μm. d Genetic deletion of SBPase by CRISPR/Cas9‐mediated genome editing. e PCR screening of a representative Δsbpase mutant. f Immunofluorescent staining confirming the loss of SBPase expression in the Δsbpase mutant. Scale bars = 5 μm. g, h Plaque assay to examine the overall fitness of the mutant. n = 3 experiments, means ± SEM; unpaired two-tailed Student’s t-test (ns, not significant, p = 0.9081). i The replication rates of the indicated strains (24 h infection, n = 3 assays, means ± SEM; ns, not significant, two-way ANOVA, p = 0.5561). j The virulence test in ICR mice. Animals were infected with 100 tachyzoites (10 mice/strain). Statistical significance was tested by log rank Mantel–Cox test (ns, not significant, p = 0.6701). Source data are provided as a Source data file.