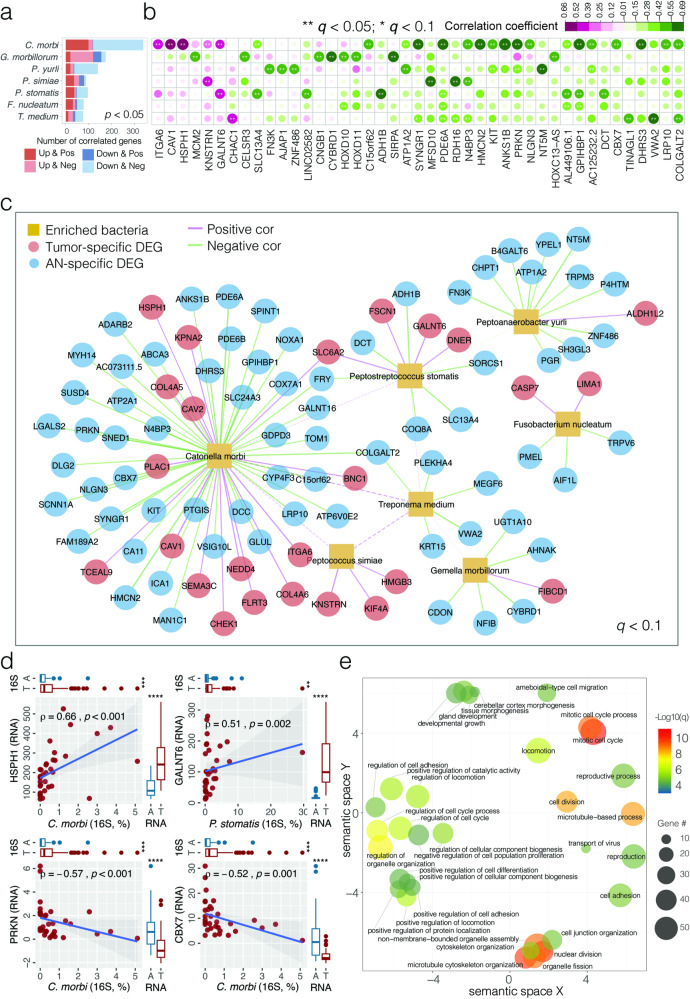
Fig. 3. Interactions between tumor-enriched bacterial species and host DEGs associated with OSCC.
a Bar plot of bacteria-transcriptome Spearman correlation showing the abundance of host DEGs in OSCC tumor tissues associated with tumor-enriched bacterial species (p < 0.05). Up and Down indicate up-regulated and down-regulated DEGs, respectively; Pos and Neg indicate positive and negative associations, respectively. C. morbi Catonella morbi, G. morbillorum Gemella morbillorum, P. yurli Peptoanaerobacter yurli, P. simiae Peptococcus simiae, P. stomatis Peptostreptococcus stomatis, F. nucleatum Fusobacterium nucleatum, T. medium Treponema medium. b Heat map depicting highly significant bacteria-transcriptome correlations (q < 0.05). Color panel indicates the Spearman correlation coefficients. Asterisks indicate significance level (**q < 0.05; *q < 0.1). Columns are ranked by the average correlation coefficients of DEGs with tumor-enriched bacteria. c Network visualizing significant correlations between bacteria-transcriptome (solid line, Spearman q < 0.1) and bacteria-bacteria (dashed line, SparCC correlation q < 0.05). d Scatter plots showing four representative correlations between bacteria and DEG in OSCC tumor tissues, where the strength of correlation (Spearman rho) and significance (p value) are shown in each plot. Marginal boxplots depict overall gene expression (right) and bacterial 16S rRNA gene abundance (top), with Mann–Whitney U test between tumor and AN tissues. e Gene Ontology (GO) pathways significantly enriched by up-regulated host DEGs associated with tumor-enriched bacteria. GO terms with functional similarity are clustered in the semantic space by REVIGO. B & H corrected p values are indicated by color panel. The size of the circles indicates the number of DEGs associated with the pathways.