Abstract
Pontocerebellar Hypoplasia (PCH) is a rare autosomal recessive hereditary neurological degenerative disease. To elaborate upon the clinical phenotypes of PCH and explore the correlation between TOE1 gene mutations and clinical phenotype, we analyze the clinical and genetic features of a Chinese infant afflicted with pontocerebellar dysplasia accompanied by gender reversal with bioinformatics methods. The main clinical features of this infant with TOE1 gene mutation included progressive lateral ventricle widening, hydrocephalus, severe postnatal growth retardation, and hypotonia, and simultaneously being accompanied by 46, XY female sex reversal. Whole exome sequencing revealed a compound heterozygous mutation in the TOE1 gene (c.299T > G, c.1414T > G), with the protein homology modeling-generated structure predicting a pathogenic variation, which is closely related to the clinical manifestations in the patient. The new mutation sites, c.299T > G and c.1414T > G, in the TOE1 gene are pathogenic variants of pontocerebellar hypoplasia type 7.
Keywords: Pontocerebellar hypoplasia, Sex reversal, TOE1 gene, Developmental delay
What is already known about this topic?
PCH is a rare autosomal recessive hereditary neurological degenerative disease that has a typical prenatal onset. Currently, very few cases of PCH type 7 have been reported, with clinical phenotype being the significant differences.
What does this study add?
We reported a cerebellopontine hypoplasia case accompanied by 46 XY female sex reversal and identified a novel compound heterozygous variant in the TOE1 gene that causes PCH7 in a Chinese child. As far as we know, this is the first case of PCH7 detected from prenatal to postnatal.
How this study might affect research, practice, or policy
Attracting more attention and clarifying the pathogenicity of the compound heterozygous variant in TOE1 gene can provide a better basis for prenatal consultation and diagnosis, enriching the PCH7 pathogenic gene spectrum for population and prenatal screening.
1. Introduction
Pontocerebellar hypoplasia (PCH) is a group of clinically diverse and genetically heterogeneous autosomal recessive neurodevelopmental disorders. Its main characteristics include delayed development and brain structural degeneration before birth [1,2]. The specific symptoms of the nervous system vary according to different subtypes. Currently, known PCH can be divided into 13 types involving 19 genes, with some types being further divided into different subtypes [3]. For example, PCH type 2, which has the highest incidence among the known subtypes, can be further divided into six subtypes, including PCH2A–PCH2F. In 2011, Anderson et al. [4] reported a full-term male infant with underdeveloped male genitalia at birth. Subsequently, penile body tissue degeneration was observed, with the genitalia surface appearing as female and having increasingly severe hypotonia, frequent apnea, and seizures. The imaging examination showed reduced white matter around the ventricles and significant pontocerebellar hypoplasia. Siriwardena et al. reported a similar case, with pontocerebellar hypoplasia having an XY sex reversal: confirmation of a new syndrome of PCH [5]. To date, pontocerebellar hypoplasia with gonadal dysplasia has been defined as PCH type 7 (PCH7, MIM#614969) [6]. However, no clear pathogenic gene was detected. Until 2017, Lardelli et al. confirmed that PCH7 is related to biallelic mutations in the 3’ exonuclease TOE1 gene [7]. Here, we reported a cerebellopontine hypoplasia case accompanied by 46 XY female sex reversal, and also detected two mutations in the TOE1 gene. Furthermore, we also analyzed the TOE1 mutation using bioinformatics methods to elucidate its role in PCH 7.
2. Methods
2.1. Object of study
An infant diagnosed with “developmental backwardness” was taken as the study subject. Their clinical data was analyzed along with imaging and genetic examinations, which included bioinformatically analyzing the TOE1 gene. This project has been approved by the hospital's Medical Ethics Committee [2022-P2-284-01]. The parents of the infant gave written informed consent.
2.2. Methods
2.2.1. Analysis of clinical characteristics
The patient's clinical phenotype and imaging results at different stages were analyzed.
2.2.2. Genetic analysis
Blood samples (2mL) were collected from the child and parents. High-throughput sequencing was performed using the Illumina NovaSeq 6000 series platform (Illumina, Inc., San Diego, CA, USA) by the Beijing FINDRARE Genetic Testing Company. The whole gene exome of patients was tested, and the sequencing data was analyzed in depth to identify possible variant genes. Comparative analysis with the human genome hg19 reference sequences (GRCh37/hg19) provided by the UCSC database was performed with Agilent's SureSelect Human All Exon V6 files and the NextGene V2.3.4 software to assess the coverage and sequencing quality of the target regions.
Variants are filtered according to strict selection criteria and annotated with relevant information from the Human Gene Mutation Database (HGMD) database and protein function prediction software. The pathogenicity of genetic variants was assessed according to the Standards and Guidelines for Interpretation of Sequence Variants published by the American College of Medical Genetics and Genomics (ACMG) in 2015, and Human Genome Variation Society (HGVS) nomenclature was used. TOE1 mRNA sequence (NM_025077.3) was chosen as the reference sequence. Both variants were detected using trio-WES and validated through Sanger sequencing.
2.2.3. Conservation analysis
The position of the PCH-related TOE1 mutation site was presented using SnapGene (https://www.snapgene.com/). TOE1 sequence comparisons between different species were analyzed using CLUSTALW (https://www.genome.jp/tools-bin/clustalw), with the synchronized TOE1 protein secondary structures analyzed using ESPript (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) [8]. The sequences being analyzed are as follows: Homo sapiens (NP_079353.3) Pan troglodytes (XP_009454561.1); Pongo abelii (NP_001125670.1); Macaca mulatta (XP_001101649.2); Equus caballus (XP_001496247.1); Sus scrofa (XP_003128093.2); Bos taurus (NP_001069062.1); Mus musculus (NP_080930.1); Canis lupus familiaris (XP_038543783.1); Loxodonta africana (XP_023410439.1); Danio rerio (NP_001243611.1); Xenopus tropicalis (NP_001120127.1).
2.2.4. Predicting the deleteriousness of mutant loci
PolyPhen-2 v2.2.3r406 (http://genetics.bwh.harvard.edu/pph2/index.shtml) [9], PROVEAN (http://provean.jcvi.org/index.php) [10] and Mutation Taster (https://www.genecascade.org/MutationTaster2021/) [11] were used to predict how an amino acid mutation possibly impacts the structure and function of a human protein using straightforward physical and comparative considerations. The Missense3D and Pymol softwares were used to analyze the potentially pathogenic mutant loci's spatial location and predict deleteriousness [12]. PredyFlexy (https://www.dsimb.inserm.fr/dsimb_tools/predyflexy/index.html) was used to predict the TOE1 sequence flexibility [13].
3. Results
3.1. Clinical characteristics analysis
General information: The infant is the second pregnancy, first delivery, born naturally at 42 gestational weeks without asphyxia at birth. The external genitalia manifested as female. The parents found the baby was unable to chase and had high muscle tension in the limbs and low muscle tension in the trunk about six weeks after birth. The baby has been confirmed to have severe neurodevelopmental and growth retardation at about one-year-old. The child is now one year and six months old with weight, length, and head circumference below the 3rd percentile and currently unable to lift their head, turn over, or chase people or objects; cortical blindness but no seizures.
3.1.1. Assistant examination
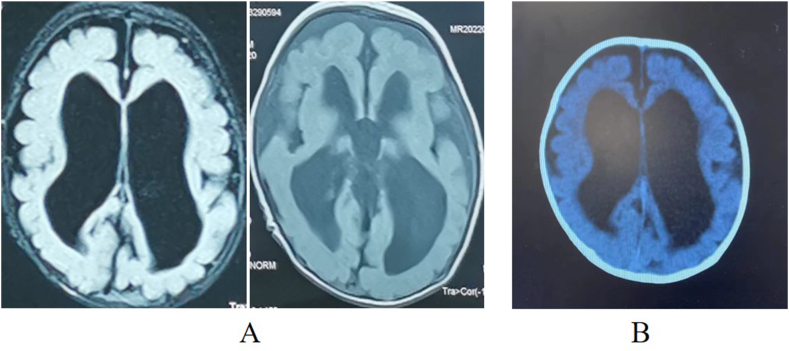
Ultrasound examination at Gestational Age (GA) 24 weeks revealed widening of the fetal lateral ventricles. Magnetic resonance imaging (MRI) at GA29 weeks revealed small volumes in both the cerebellar hemispheres, wide cerebellomedullary cisterns, slender corpus callosum, and widened bilateral ventricles. Postnatal brain image suggests severe hydrocephalus. (Fig. 1A-B).
Fig. 1.
Brain imaging at different stages. (A) Head MRI 1 month post-birth. (B) Head CT at 3 months post-birth. All indicate severe hydrocephalus.
3.1.2. Growth development assessment
If the body weight/length/head circumference is lower than the 10th percentile (p10) of the same age, it is considered a developmental delay. In contrast, if it is lower than the 3rd percentile (p3), it is considered a severe developmental delay. The baby had a birth weight of 3.4 kg (p50), length of 49 cm (p25), head circumference of 33.5 cm (p15). Six months after birth: the weight was 5.5 kg (below p3), length was 65 cm (p10), and head circumference was 38.3 cm (below p3). One year after birth: the weight was 5.5 kg (below p3), length was 65 cm (below p3), and head circumference was 40 cm (below p3). At birth, the infant is appropriate for gestational age and experiences progressive growth delay. Growth development assessment was performed according to the WHO Child Growth Standards (http://www.who.int/child growth/en).
3.1.3. Neurodevelopment assessment
Neurodevelopment assessment was administered at three months and six months old, respectively. Here, five major functional areas, including adaptive, gross motor, fine motor skills, and language and personal-social areas, were assessed by the Gesell scale and they showed progressive regression (Suppl. Table 1).
3.1.4. Electroencephalogram of the child (5 months after birth)
The background showed a moderate to high amplitude 34 Hz rhythm or non-rhythmic slow waves, mixed with a small amount of low amplitude θ Wave. We observed no spindle wave during sleep, a visible apex wave, and no epileptiform discharge. There was incomplete inhibition of the occipital rhythm.
3.1.5. Chromosome examination of the father
46, XY, inv(9) (p12q13)with an arm-to-arm inversion in chromosome 9.
3.1.6. Chromosome examination of the mother
46, XX, with no abnormal conditions.
3.1.7. Fetal chromosome examination (amniotic fluid sample)
arr(1–22) × 2,(X,N) × 1. There was no clinically significant copy number variation found within the detection range.
3.1.8. Chromosome karyotype of the infant
46, XY.
3.1.9. Maternal pregnancy history
Gravida 2, para 1, routine pregnancy examination at 24 weeks of the first pregnancy, found that the fetal lateral ventricle had widened and had hydrocephalus, with induced abortion being performed later. However, no relevant inspections were conducted. There were no complications during this pregnancy, with prenatal examinations being regularly conducted. Fetal abnormalities were also found at 24 weeks of pregnancy.
3.2. Genetic analysis and sanger sequencing verification of the TOE1 gene
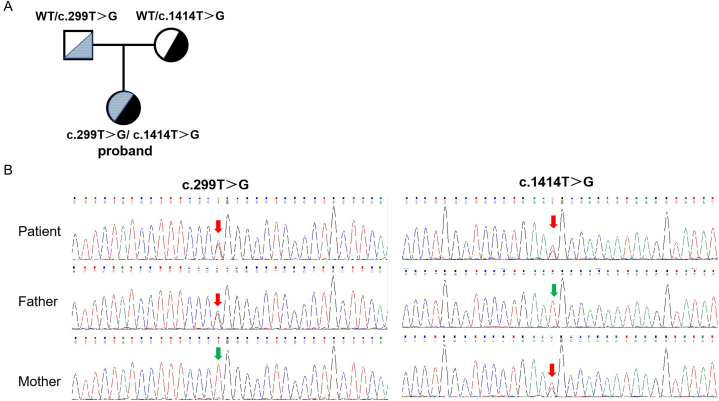
An inherited compound heterozygous variant, NM_025077.3: c.299T > G (p.L100R) and c.1414T > G (p.C472G), was found in the TOE1 gene of the child. The mutation sites were validated by Sanger sequencing. The proband's father carried the c.299T > G (p.L100R) heterozygous variant, and the mother carried the c.1414T > G (p.C472G) heterozygous variant. (Fig. 2A-B). These two loci have not been previously reported.
Fig. 2.
Clinical and molecular characteristics of the patient. (A) Family pedigree of the proband. (B) Sanger sequencing revealed that the patient harbored a compound heterozygous variant (c.299T > G and c.1414T > G, p.L100R and p.C472G) in the TOE1 gene (NM_025077.4). Red arrows indicate the variant base.
3.3. Structural analysis of novel compound heterozygous mutations
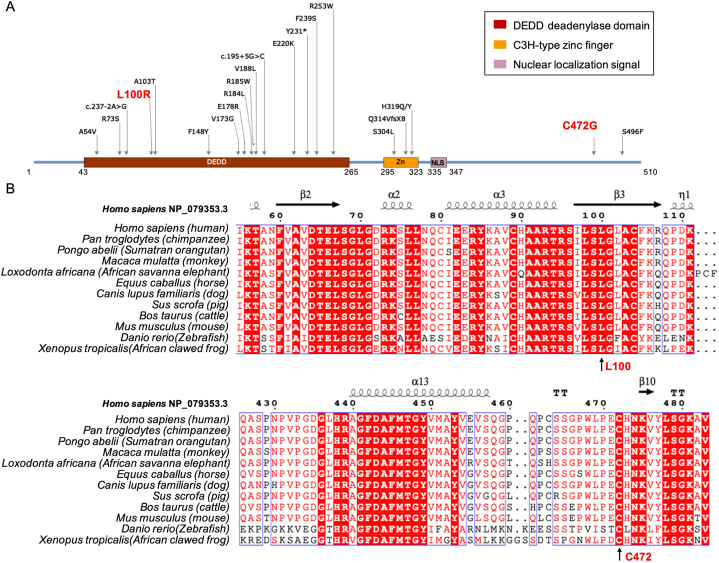
We demonstrated the novel TOE1 gene mutation sites, i.e., c.299T > G (p.L100R) and c.1414T > G (p.C472G), along with the reported PCH-related mutation sites on the TOE1 protein. Here, p.L100R was located in the Asp-Glu-Asp-Asp (DEDD) deadenylase domain, whereas p.C472G was located in the carboxyl end of amino acid (Fig. 3A). Conservation analysis indicated Leu 100 and Cys 472 are highly conserved among the various species (Fig. 3B).
Fig. 3.
Novel compound heterozygous mutations in TOE1 gene. (A) p.L100R was located in the DEDD domain, whereas p.C472G was located in the carboxyl end of amino acid. (B) Interspecies amino acid sequence alignment of the TOE1 gene across the different vertebrate species.
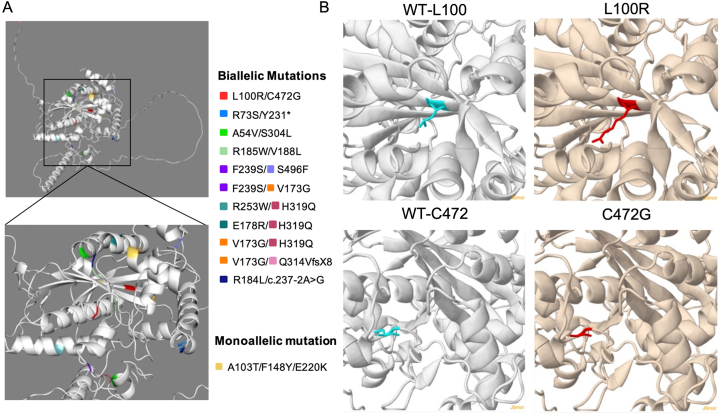
We demonstrated the spatial location of the newly found mutation site in this study along with the other previously reported mutations on the 3D model of the TOE1 protein, with the new mutations found in this study (red) shown as a stick model (Fig. 4A and B). The 3D structures of the molecule and missense variant were analyzed using the Missense3D software. The p.L100R substitution does not trigger a clash alert, which will be triggered when the mutant structure has a MolProbity clash score ≥30 and the increase in clash score is > 18 compared to the wild type [14]. The wild-type and mutant local clash scores were 15.29 and 19.26, respectively. However, this mutation does not alter the secondary structure ‘E’ (extended strand in parallel and/or anti-parallel β-sheet conformation). However, this substitution replaces a buried hydrophobic and uncharged residue (Leu, relative solvent accessibility, RSA, 4.2%) with a hydrophilic and charged residue (Arg, RSA 0.0%). Furthermore, the p.C472G substitution caused a switch between the buried and exposed states of the target variant residue. Here, the Cys was buried (RSA 0.7%), whereas Gly was exposed (RSA 11.9%). However, this substitution does not alter the secondary structure ‘T’ (hydrogen-bonded turn). Although the wild-type residue was Cys, it did not form a disulfide bond with any neighboring wild-type residue. Thus, no disulfide bonding pattern changed in the mutation sequence (Fig. 4B).
Fig. 4.
Spatial location of the TOE1 gene mutation site. (A) the spatial location of the newly found mutation in this study with reported mutations. (B) the spatial location of the TOE1 gene mutation on the wild-type model of TOE1 protein, with the mutation site being shown as the stick model, and the new mutations found in this study are shown in red.
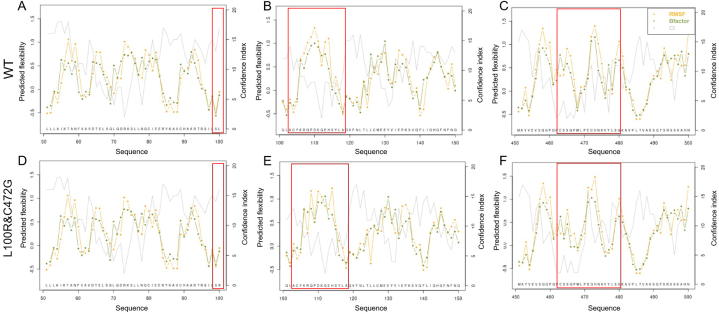
Since the p.L100R and p.C472G mutations did not affect the secondary structure of the TOE1 protein, we proceeded to analyze the effect of these two mutation sites on the flexible structure of the TOE1 protein. The B-factor was used to describe the thermal motion-induced attenuation of X-ray or neutron scattering and the protein flexibility. The higher the B-factor in the protein structure, the better the motility, and conversely, a lower B-factor meant that the structure was more rigid [15]. Normalized Root Mean Square Fluctuations (RMSF) values represent the degree of freedom of movement of individual atoms in a molecule. They can characterize the flexibility and thermal stability of the protein conformation [16]. The results showed that biallelic mutations (p.L100R and p.C472G) change the flexible structure between Leu 100 to Ala 118, which is located in the DEDD deadenylase domain, and Cys 463 to Gly 480 (Fig. 5A–F). Therefore, these results indicated that p.L100R and p.C472G might confer flexibility and thermal stability to the TOE1 protein.
Fig. 5.
Flexible structure prediction of the TOE1 protein along sequence. (A–C) flexible structure of wild-type TOE1 protein, (D–F) flexible structure of mutated TOE1 protein (p.L100R and p.C472G). Each line point represents a particular measure. Green and orange lines represent the B-factor and RMSF prediction, respectively, which were deduced from the flexibility class prediction. The gray line represents the confidence index. The higher was the confidence index, the more accurate the prediction was thought to be.
3.4. Pathogenicity analysis of novel compound heterozygous mutations
The results of the pathogenicity analysis according to the ACMG showed that c.299T > G (p.L100R) and c.1414T > G (p.C472G) variants were not reported in the literature. Bioinformatics analysis software (PolyPhen2, PROVEAN and Mutation Taster) predicted deleterious outcomes (PP3) of these variants. PolyPhen-2 predicted that the p.L100R variant would impair the TOE1 function, according to HumDiv (“probably damaging,” score = 1) and HumVar (“probably damaging,” score = 0.998). The p.C472G variant showed “probably damaging” with scores of 0.998 and 0.897, respectively. The score of PolyPhen-2 ranges from zero to one; the larger the score, the more harmful it was. The PROVEAN score of p.L100R and p.C472G were −4.975 and −6.141, respectively, which were considered “deleterious” (cutoff = −2.5). The analysis on Mutation Taster came to the same conclusion. Therefore, these results indicated that p.L100R and p.C472G might damage the function of TOE1. These variants were not included in the normal population database gnomAD (PM2_Supporting). Based on current evidence, the variant was defined as a variation of undetermined clinical significance (PM2_Supporting + PP3).
4. Discussion
PCH is a rare autosomal recessive hereditary neurological degenerative disease that has a typical prenatal onset [17]. The common features of PCH include pons and cerebellum atrophy, progressive microcephaly, different degrees of ventricular dilation, severe cognitive impairment, motor disorders, and seizures. Lardelli et al. [7] reported 12 PCH7 families and found that most families had biallelic gene function deletion mutations in TOE1 (NC_000001.11), with further animal models and functional verification proving that these mutations can reduce and cause abnormality in the TOE1 functionality. Unfortunately, it is unclear how this affects the development of the pons, cerebellum, and testes in the fetal period.
Currently, very few cases of PCH7 have been reported, with clinical phenotype being the significant difference. Only 16 cases of PCH 7 have been reported worldwide, with four found in China among two siblings from one family [[18], [19], [20]]. Here, we reported, we found the hydrocephalus, widened medullary cistern, cerebellar hypoplasia, and corpus callosum hypoplasia starting from the fetal period; as far as we know, this is the first case of PCH7 detected from the prenatal to postnatal stages. The neuroimaging was like the typical PCH described previously by Rüsch [21]. Furthermore, we also found that postnatal neurological development was severely impaired, along with severely delayed growth development. However, there was no significant respiratory arrest or epileptic seizures. Based on the results of genetic testing, for the TOE1 compound heterozygous mutation, the primary diagnosis was pontocerebellar dysplasia type 7. Since this was mostly accompanied by gonadal dysplasia, we conducted chromosomal karyotype analysis for this infant, and the results confirmed that the patient is 46, XY. The infant's external genitalia was female, but there were no testicles, ovaries, or uterus in its pelvic cavity. These two compound heterozygous mutations in TOE1 have not been reported yet. Therefore, our findings will broaden the phenotype spectrum of neurodegenerative diseases associated with TOE1 mutations.
The TOE1 gene encodes a nuclear adenylase comprising a C3H zinc finger (ZN), a nuclear localization signal (NLS) peptide, and a DEDD domain with deadenylase activity. Its function was to inhibit cell growth and cycle, mediate the growth inhibition effect of EGR1, and participate in the maturation and the processing of the 3′ tail of snRNA [22]. Although the biallelic mutations of TOE1 form the foundation of PCH7, there are currently scattered case reports of the TOE1 compound heterozygous mutations leading to severe phenotypes, with the mechanism still remaining unknown.
Bioinformatics analysis indicated that c.299T > G p.L100R and c.1414T > G p.C472G might damage TOE1 function. Interestingly, the p.L100R variants of our patient are in the DEDD domain and affected the surface residues rather than the β-sheet conformation. Correspondingly, the B-value and RMSF value between Leu 100 and Ala 118 had changed. The p.C472G mutation resulted in the B-value and RMSF value fluctuating in the zone from Cys 463 to Gly 480. The catalytic activity center of proteases is usually located in the low B-value region, and this feature was used to predict the location of the enzyme activity center. Later, it was found that protein thermal stability was closely related to its rigidity. Thus, the variants were likely to affect the deadenylase activity directly.
Knockdown of TOE1 was found to cause developmental arrest during the morula-to-blastocyst transition in mice [23]. This developmental arrest can be rescued by TOE1 mRNA microinjection, which indicates that TOE1 plays an important role in early embryonic development. However, whether TOE1 can serve as an effective therapeutic target for PCH7 requires further in-depth studies. In conclusion, our study identified one compound heterozygous variant in the TOE1 gene, enriching the PCH7 pathogenic gene spectrum for population and prenatal screening.
Ethics approval and consent to participate
This project has been approved by the hospital's Medical Ethics Committee [2022-P2-284-01].
Consent for publication
All authors consent for publication.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Funding
This study was supported by Special Projects on health development in the capital(No.2022-1-2111).
CRediT authorship contribution statement
Tianli Wei: Writing – original draft, Investigation. Shuguang Shan: Investigation. Zhaojun Jia: Writing – review & editing, Methodology. Yingxue Ding: Supervision, Project administration, Methodology, Investigation, Data curation.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Yingxue Ding reports writing assistance was provided by EditSprings.com. No reports a relationship with No that includes:. No has patent pending to No. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the patient and parents participated in this study and all clinicians involved in the case discussion. The authors would like to express their gratitude to EditSprings (https://www.editsprings.com/) for the expert linguistic services provided.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28678.
Contributor Information
Tianli Wei, Email: weitl0830@163.com.
Shuguang Shan, Email: jiute2020@163.com.
Zhaojun Jia, Email: jiazj@bipt.edu.cn.
Yingxue Ding, Email: d-yingxue@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rüsch C.T., Bölsterli B.K., Kottke R., Steinfeld R., Boltshauser E. Pontocerebellar hypoplasia: a pattern recognition approach. Cerebellum. 2020;19(4):569–582. doi: 10.1007/s12311-020-01135-5. [DOI] [PubMed] [Google Scholar]
- 2.van Dijk T., Barth P., Baas F., Reneman L., Poll-The B.T. Postnatal brain growth patterns in pontocerebellar hypoplasia. Neuropediatrics. 2021;52(3):163–169. doi: 10.1055/s-0040-1716900. [DOI] [PubMed] [Google Scholar]
- 3.Bilge S., Mert G.G., Hergüner Ö., et al. Ital J Pediatr. Clinical, radiological, and genetic variation in pontocerebellar hypoplasia disorder and our clinical experience. 2022;48(1):169. doi: 10.1186/s13052-022-01349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson C., Davies J.H., Lamont L., Foulds N. Early pontocerebellar hypoplasia with vanishing testes: a new syndrome? Am. J. Med. Genet. 2011;155A(4):667–672. doi: 10.1002/ajmg.a.33897. [DOI] [PubMed] [Google Scholar]
- 5.Siriwardena K., Al-Maawali A., Guerin A., et al. XY sex reversal, pontocerebellar hypoplasia and intellectual disability: confirmation of a new syndrome. Am. J. Med. Genet. 2013;161A(7):1714–1717. doi: 10.1002/ajmg.a.35945. [DOI] [PubMed] [Google Scholar]
- 6.Van Dijk T., Baas F., Barth P.G., Poll-The B.T. What's new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet J. Rare Dis. 2018;13(1):92. doi: 10.1186/s13023-018-0826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lardelli R.M., Schaffer A.E., Eggens V.R., et al. Biallelic mutations in the 3' exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat. Genet. 2017;49(3):457–464. doi: 10.1038/ng.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinhaus R., Proft S., Schuelke M., et al. MutationTaster2021. Nucleic Acids Res. 2021;49(W1):W446–W451. doi: 10.1093/nar/gkab266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ittisoponpisan S., Islam S., Khanna A., et al. Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? J. Mol. Biol. 2019;431(11):2197–2212. doi: 10.1016/j.jmb.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Brevern A.G., Bornot A., Craveur P., Etchebest C., Gelly J.C., et al. PredyFlexy: flexibility and local structure prediction from sequence. Nucleic Acids Res. 2012;40(Web Server issue):W317–W322. doi: 10.1093/nar/gks482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen V.B., Arendall W.B., Headd J.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z., Liu Q., Qu G., Feng Y., Reetz M.T. Utility of B-factors in protein science: interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem. Rev. 2019;119(3):1626–1665. doi: 10.1021/acs.chemrev.8b00290. [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Wei B., Che C., et al. Enhanced stability of manganese superoxide dismutase by amino acid replacement designed via molecular dynamics simulation. Int. J. Biol. Macromol. 2019;128:297–303. doi: 10.1016/j.ijbiomac.2019.01.126. [DOI] [PubMed] [Google Scholar]
- 17.Zakaria R.B.M., Malta M., Pelletier F., et al. Classic "PCH" genes are a rare cause of radiologic pontocerebellar hypoplasia. Cerebellum. 2023 Mar 27 doi: 10.1007/s12311-023-01544-2. [DOI] [PubMed] [Google Scholar]
- 18.Chen H., Li N., Xu Y., et al. Novel compound heterozygous variant of TOE1 results in a mild type of pontocerebellar hypoplasia type 7: an expansion of the clinical phenotype. Neurogenetics. 2022;23(1):11–17. doi: 10.1007/s10048-021-00675-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang C., Ge Y., Li R., et al. Novel compound heterozygous missense variants in TOE1 gene associated with pontocerebellar hypoplasia type 7. Gene. 2023;862 doi: 10.1016/j.gene.2023.147250. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z.F., Lv K.L., Yao S.Q., et al. Clinical and genetic characterization of Chinese family with pontocerebellar hypoplasia type 7. Am. J. Med. Genet. 2024;194(1):46–52. doi: 10.1002/ajmg.a.63371. [DOI] [PubMed] [Google Scholar]
- 21.Kasinathan A., Sankhyan N., Dijk T.V., et al. Clinico-radiological profile of children with pontocerebellar hypoplasia. J. Pediatr. Neurosci. 2020;15(2):94–98. doi: 10.4103/jpn.JPN_6_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng T., Huang Y., Weng K., et al. TOE1 acts as a 3' exonuclease for telomerase RNA and regulates telomere maintenance. Nucleic Acids Res. 2019;47(1):391–405. doi: 10.1093/nar/gky1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Ming X., Zhang S., et al. Knockdown of Toe1 causes developmental arrest during the morula-to-blastocyst transition in mice. Theriogenology. 2022;194:154–161. doi: 10.1016/j.theriogenology.2022.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.