Abstract
Krabbe disease is a sphingolipidosis characterized by the genetic deficiency of the acid hydrolase β-galactosylceramidase (GALC). Most of the studies concerning the biological role of GALC performed on Krabbe patients and Galc-deficient twitcher mice (an authentic animal model of the disease) indicate that the pathogenesis of this disorder is the consequence of the accumulation of the neurotoxic GALC substrate β-galactosylsphingosine (psychosine), ignoring the possibility that this enzyme may exert a wider biological impact. Indeed, limited information is available about the effect of GALC downregulation on the cell lipidome in adult and developing organisms. The teleost zebrafish (Danio rerio) has emerged as a useful platform to model human genetic diseases, including sphingolipidoses, and two GALC co-orthologs have been identified in zebrafish (galca and galcb). Here, we investigated the effect of the competitive and irreversible GALC inhibitor β-galactose-cyclophellitol (GCP) on the lipid profile of zebrafish embryos. Molecular modelling indicates that GCP can be sequestered in the catalytic site of the enzyme and covalently binds human GALC, and the zebrafish Galca and Galcb proteins in a similar manner. Accordingly, GCP inhibits the β-galactosylceramide hydrolase activity of zebrafish in vitro and in vivo, leading to significant alterations of the lipidome of zebrafish embryos. These results indicate that the lack of GALC activity deeply affects the lipidome during the early stages of embryonic development, and thereby provide insights into the pathogenesis of Krabbe disease.
Keywords: Lipidomics, Galactosylceramidase, Krabbe disease, Molecular modeling, Zebrafish
Graphical Abstract
1. Introduction
Krabbe disease, also known as globoid cell leukodystrophy (OMIM #245200), is an autosomal recessive sphingolipidoses characterized by the deficiency of the acid hydrolase β-galactosylceramidase (GALC) encoded by the GALC gene. GALC catalyzes the removal of β-galactose from β-galactosylceramide, a major component of myelin, and other terminal β-Gal-containing sphingolipids [1]. The early infantile form of the disease is characterized by fast progression and early death. The symptoms include irritability, regression of psychomotor development, feeding difficulties, followed by hypertonicity, seizures, and loss of vision and hearing [2]. Hematopoietic stem cell transplantation is the standard of care of Krabbe disease. However, it improves the lifespan of Krabbe patients only when performed before symptoms outbreak [3].
Based on the recently confirmed “psychosine hypothesis” [4], Krabbe disease is the consequence of the accumulation of the neurotoxic GALC substrate β-galactosylsphingosine (psychosine) in the central and peripheral nervous system. This leads to neuroinflammation, degeneration of oligodendroglia, and progressive demyelination [5]. In this context, most of the studies performed on Krabbe patients and Galc-deficient twitcher mice [an authentic animal model of the disease [6]] have led to the idea that the major biological function of GALC may be its psychosine “scavenging” activity, and the possibility that this enzyme may exert a wider biological impact has been neglected.
The teleost zebrafish (Danio rerio) represents a useful platform to model human genetic diseases. In addition, lipidomic studies performed in zebrafish have identified all the main classes of lipids present in mammals, supporting the possibility to model diseases of the lipidic metabolism in this animal species, including sphingolipidoses [7].
Two GALC co-orthologs have been identified in zebrafish (galca and galcb). They share a high level of sequence identity with their human counterpart and are co-expressed in the central nervous system during embryonic development [8]. Double galca/galcb knockdown by oligonucleotide morpholino injection caused alterations of the central nervous system in the absence of a significant accumulation of psychosine, suggesting that the loss of GALC may have pathological consequences during embryonic development that are independent of psychosine accumulation [8]. Accordingly, alterations of the myelinated regions of the spinal cord, brain stem, and peripheral nerves have been observed in 20–23-week-old Krabbe fetuses, before a significant accumulation of psychosine can occur [9], [10]. Nevertheless, no data are available about the impact of GALC deficiency on the lipid profile of developing organisms.
The intrauterine gestation makes it difficult to follow the embryonic alterations that may occur in murine models of Krabbe disease. By contrast, the rapid in vitro development as optically transparent embryos and the fact that all major organ systems are established within 5 days post-fertilization make zebrafish suitable for the study of the impact of GALC deficiency on the lipid landscape during embryonic development. Here, we have investigated the effect of the competitive and irreversible activity-based GALC inhibitor Gal-cyclophellitol (GCP) [11] on the lipid profile of zebrafish embryos. Computational docking and molecular dynamics (MD) simulations show the ability of GCP to sequester in the catalytic site and covalently bind the human GALC and the zebrafish orthologues. Accordingly, GCP inhibits the β-galactosylceramide hydrolase activity of zebrafish in vitro and in vivo, leading to significant alterations of the lipid profile of zebrafish embryos.
These results indicate that the lack of GALC activity may affect the lipidome during the early stages of embryonic development, providing insights into the pathogenesis of Krabbe disease and setting the basis for improving the comprehension of the alterations of the central and peripheral nervous system observed in human Krabbe fetuses.
2. Materials and methods
2.1. Molecular docking studies
Human GALC (hGALC) protein (Uniprot P54803) was modelled on the Swiss-Model website (https://swissmodel.expasy.org/) [12] based on the murine GALC structure with β-D-galactose (β-D-Gal) bound determined by x-ray crystallography (PDBid: 4CCE: sequence identity 82.6%) [13]. Similarly, the initial models of the zebrafish Galca (Uniprot Q5SNX7) and Galcb (Uniprot Q7ZUD5) proteins were modelled on the Swiss-Model website based on the murine GALC/saposin-A crystal structure (PDBid: 5NXB; sequence identity 64.48% and 64.99%, respectively) [14]. The high sequence identity and the conservation of the catalytic site enabled the generation of high-quality models as evaluated by the MolProbity tool [15]. The protein structures were then prepared using the Protein Preparation Wizard in Maestro (Schödinger Release 2019–1) [16] and protonated at pH 4.4 using PROPKA. Prior to the docking calculation, β-D-Gal and GCP were prepared with LigPrep [16] and ionized at pH 4.4 using Epik [17]. Flexible non-covalent docking (hereafter referred to as flexible docking) studies were performed with Glide 4.8 [16] in Standard Precision mode with default parameters at pH 4.4. The grid box was centered on the active site pocket: G64, T109, N197, E198, E274, W307, Y319, and W541 for human GALC (UniProt P54803); G41, T87, N175, E176, E251, W284, F296, and W517 for zebrafish Galca (Uniprot Q5SNX7); G45, T91, N179, E180, E256, W289, F301, and W521 for zebrafish Galcb (Uniprot Q7ZUD5). Notably, E198 in human GALC, and E176 and E180 in zebrafish Galca and Galcb, respectively, were protonated. Covalent docking was performed with Glide.4.8 [16] in covalent docking mode with default parameters at pH 4.4. The grid box was centered as previously described. The reaction type selected was the epoxide opening while the reacting residue selected was E274 in human GALC and E251 and E256 in zebrafish Galca and Galcb, respectively [13]. Thirty-two poses for each ligand were collected and ranked according to their Gscore value for β-D-Gal or GCP with human GALC or the zebrafish orthologues in the flexible or covalent docking procedures. The poses selected for β-D-Gal and GCP in both the flexible docking and the covalent docking were the top-ranked poses, which overlapped with the orientation of β-D-Gal in the crystal structure (PDBid: 4CCE) [13].
2.2. All-atom molecular dynamics (MD) simulation
The Amber18 package [18] was used to carry out the simulations for the modelled systems starting from the top-ranked docking poses. Three replica all-atom MD simulations of 100 ns duration in explicit solvent and 150 mM NaCl salt concentration were run for each system with either β-D-Gal or GCP bound non-covalently. Parameters for GALC were assigned with the ff14SB force field [19]. Ligands were parameterized with GAFF [20] along with AM1-BCC for assigning partial charges [21]. The models were placed in a periodic-cubic water box using the TIP3P water model [22] with 10 Å between the solutes and the edges of the box. Na+ and Cl- ions were added to neutralize the systems and to immerse them in solvent with an ionic strength of 150 mM. Each system was energy minimized in 4 consecutive minimization steps, each of 100 steps of steepest descent followed by 900 steps of conjugate gradient, with decreasing positional restraints from 10 to 0 kcal/mol Å−2 on all the atoms of the systems excluding waters, counterions, and hydrogens, with a cut-off for non-bonded interactions of 8 Å. The systems were then subjected to two consecutive steps of heating, each of 10,000 steps, from 10 to 100 K and from 100 to 310 K in an NVT ensemble with a Langevin thermostat. Bonds involving hydrogen atoms were constrained with the SHAKE algorithm and 2 fs time step was used. The systems were then equilibrated at 310 K for 2.5 ns in the NPT ensemble with a Langevin thermostat with random velocities assigned at the beginning of each step. During the MD simulations, a cutoff of 8 Å for the evaluation of short-range non-bonded interactions was used and the Particle Mesh Ewald method was employed for the long-range electrostatic interactions. The temperature was kept constant at 310 K with a Langevin thermostat. MD trajectories were analyzed using CPPTRAJ from AmberTools20 [18] and Visual Molecular Dynamics (VMD) [23]. The binding free energies (∆Gtotal) and their components were calculated with the Molecular Mechanics Generalized Born Surface Area (MM-GBSA) method by using the MMPBSA.py program from AmberTools20 [18] on 50 frames taken from the equilibrated part of the trajectories (10 - 100 ns). The entropic contribution to the free energy was not considered, as this term does not generally improve the quality of the results using this method [24]. Moreover, a per-residue decomposition analysis was carried out.
2.3. Zebrafish maintenance
Zebrafish embryos were handled according to relevant national and international guidelines. Current Italian rules do not require approval for research on zebrafish embryos. Zebrafish were raised and maintained under standard laboratory conditions as described [25]. Briefly, the wildtype AB strain and the transgenic reporter Tg(neurod1:EGFP)ia50 line [26] were maintained at 28°C on a 14 h light/10 h dark cycle. Immediately after spawning, the fertilized eggs were harvested, washed, and placed in 10 cm Petri dishes in fish water. Embryos were incubated at 28 °C and staged as described [27].
2.4. GALC activity assay
GALC activity was evaluated in the extracts of zebrafish embryos or of adult zebrafish or mouse brains. For this purpose, samples were sonicated in 0.25 M sucrose/1.0 mM EDTA, pH 7.2, in the presence of a cocktail of protease inhibitors. After evaluation of protein concentration, GALC-mediated hydrolysis of the fluorescent GALC substrate lissamine-rhodaminyl-6-aminohexanoyl-galactosylceramide (LRh-6-GalCer) was quantified by thin-layer chromatography (TLC) [28] following incubation of the substrate with 20–50 μg of the various tissue extracts. Briefly, 3 nmoles of LRh-6-GalCer in 3:2 chloroform/methanol were concentrated and dissolved in 5 μl of DMSO and 25 μl of 0.2 M citrate phosphate buffer, pH 4.4. The extracts and water were added to a final volume of 100 μl and incubated overnight at 37 °C. Next, the reaction was added with 1.9 ml of 3:2 v/v chloroform/methanol and 0.4 ml of water. After vortexing, the organic phase was separated by centrifugation, collected, and evaporated under nitrogen. Samples dissolved in 3:2 v/v chloroform/methanol were spotted on glass-coated silica gel plates and developed in 25:25:25:9:16 volumes chloroform/ethyl acetate/n-propanol/0.25 M KCl/methanol. The fluorescent ceramide spots (LRh-6-Cer) were visualized under an ultraviolet lamp and photographed. Next, the bands corresponding to the enzyme product were extracted in 3:2 volumes chloroform/methanol and fluorescence of the solubilized product was measured at the spectrofluorometer (Excitation: 565 nm; Emission: 575 nm).
2.5. Light sheet microscopy
Tg(neurod1:EGFP)ia50 zebrafish embryos at shield stage (6 hpf) were injected with vehicle (4 nanoliters of 10% DMSO in water) or with the maximal tolerated dose of GCP in vehicle (160 pmoles/embryo) as assessed by a series of preliminary experiments (data not shown). Then, embryos were first anesthetized using 0.16 mg/ml tricaine, embedded in 0.5% low melting agarose (Top Vision Low Melting Point Agarose, Thermo Fisher Scientific), and mounted on glass capillaries (Carl Zeiss, Germany). Images were acquired using Zeiss LightSheet microscope V1 supported by ZenPro software. EGFP acquisition was performed using 488–30 nm laser and 505–545 nm filter. Images from the same experiment were taken with the same laser intensity and exposure time to generate comparable images. After acquisition, 3D images were generated using Arivis software (Zeiss). 3D reconstructions were exported as a single snap with the same compression settings.
2.6. LC-MS lipidomic analysis of zebrafish embryos
Zebrafish embryos at 1–2 cell stage were injected with vehicle (4 nanoliters of 10% DMSO in water) or with GCP in vehicle (160 pmoles/embryo). At 96 h post-fertilization (hpf), embryos were harvested and grouped in 3 pools of GCP or vehicle treaded animals, each formed by 4–8 embryos. Next, pools were processed for GALC activity assay and lipidomic analysis. Lipids were extracted using two different cold organic solvents. Embryos were sonicated, and 5 μl of a methanol mix of deuterated standards (Splash Lipidomix®) was added. Then, 225 μl of cold methanol were added and the sample was vortexed for 10 s, followed by the addition of 750 μl of cold methyl-tert-butyl ether and vortexed for 10 s. The samples were then shacked at 4 °C for 6 min at 3000 rcf and 100 μl of water were added and vortexed. After 2 min of centrifugation at 20800 rcf and 4 °C, 500 μl of supernatant were collected and evaporated using a SpeedVac. The dried sample was reconstituted with 50 μl of a solution MeOH/toluene 9:1 (v/v) containing the internal standard CUDA (12.5 ng/ml). For the lipidomic analysis a UHPLC Vanquish system (Thermo Scientific, Rodano, Italy) coupled with an Orbitrap Q-Exactive Plus (Thermo Scientific, Rodano, Italy) was used. A reverse phase column was used for the separation of lipids (Hypersil GoldTM 150 ×2.1 mm, particle size 1.9 µm). Mass spectrometry analysis was performed in both positive and negative ion modes. The source voltage was maintained at 3.5 kV in the positive ion mode and 2.8 kV in the negative ion mode. All other interface settings were identical for the two types of analysis. The injection volume was 3 μl. Lockmass and regular inter-run calibrations were used for accurate mass-based analysis. An exclusion list for background ions was generated by analyzing the same procedural blank sample, for both the positive and negative ESI modes. More details on the chromatographic and mass spectrometry conditions can be found here [29]. To ensure good reproducibility during the analysis, different quality control procedures were adopted: a quality control sample, represented by a pool of all the samples, was analyzed at the beginning of the batch, after every 5 samples, and at the end of the batch. Internal standards that cover several analyte classes at appropriate levels (Avanti SPLASH Lipidomix), and an internal standard (CUDA) added before the LC-MS analysis were used. Instrument variations were then monitored and corrected ensuring the good reproducibility of all the batches. Raw data acquired from untargeted analysis were processed with MSDIAL software (Yokohama City, Kanagawa, Japan), version 4.24. Peaks were detected, MS2 data were deconvoluted, compounds were identified, and peaks were aligned across all samples. For quantification, the peak areas for the different molecular species detected were normalized using the deuterated internal standard for each lipid class. To obtain an estimated concentration expressed in nmol/4 embryos, the normalized areas were multiplied by the concentration of the internal standard. An in-house library of standards was also used for lipid identification. MetaboAnalyst 4.0 software (www.metaboanalyst.org) was used for the statistical analysis.
2.7. Statistical analysis
Statistics were performed using Microsoft Excel 365 and Graphpad Prism 8. P-values were calculated by a two-tailed uncoupled t-test and P-values ≤ 0.10 were considered statistically significant.
3. Results
3.1. MD simulations and covalent docking elucidate the activity of the GCP inhibitor on human GALC
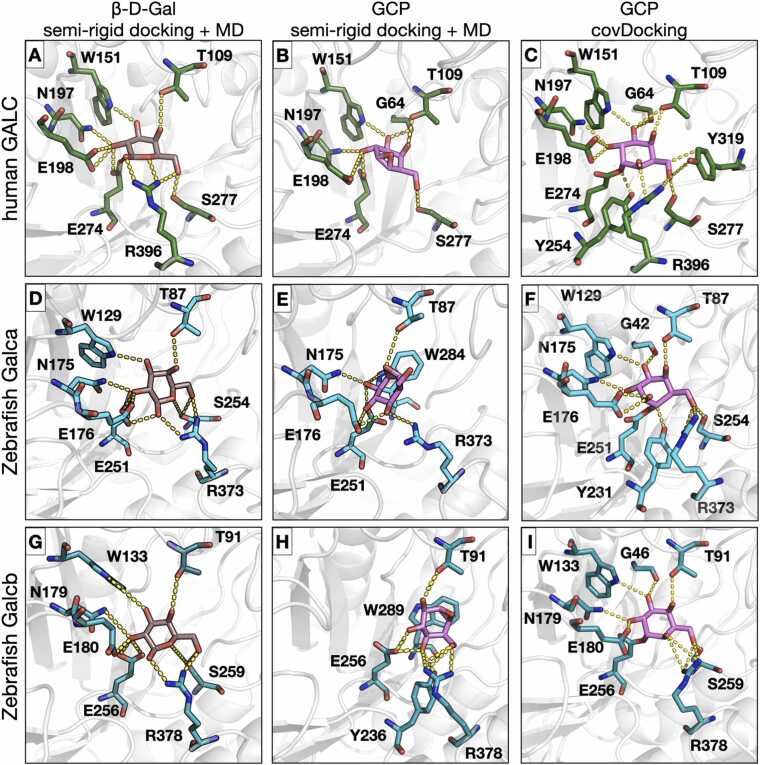
To investigate the effects of the selective inhibitor GCP, the structure of human GALC (hereafter named hGALC) was modelled by homology using the Swiss-Model webserver [12] with the murine GALC crystal structure (PDBid: 4CCE) [13] as a template. The global amino acid sequence identity between the two proteins is 82.6% and the local identity in the catalytic site increases up to 100% (UniProt P54818 and P54803 for murine and human sequences, respectively). The binding mode of the natural product β-D-Gal and of the irreversible inhibitor GCP in hGALC was investigated by performing flexible docking with the Glide docking software [16] at pH 4.4, in keeping with the lysosomal pH [30]. The top-ranked poses for both ligands perfectly overlapped with the binding mode of β-D-Gal in the mouse GALC crystal structure. Indeed, the root-mean-square deviation (RMSD) computed for β-D-Gal in the mouse and human proteins was equal to 0.1 Å and between the heavy atoms of β-D-Gal and GCP in the human enzyme was equal to 0.5 Å. This indicates that the catalytic pocket is conserved between the murine and human enzymes and points to a similar binding mode for β-D-Gal and GCP. β-D-Gal establishes hydrogen bonds with T109, W151, N197, E198, E274, S277, and R396 residues of human GALC, while the non-covalently bound GCP interacts with G63, T109, W151, N197, E198, E274, and S277 residues (Table 1). The docking scores indicate a slightly better binding affinity for GCP (Gscore= −7.85) when compared to β-D-Gal (Gscore= −7.35).
Table 1.
Docking scores and amino acid residues of human GALC and zebrafish orthologues involved in interaction with β-D-Gal and GCP in the docked complexes.
| Enzyme | Ligand | Gscore | RMSD (Å) | Amino acid residues |
|---|---|---|---|---|
| hGALC | β-D-Gal | -7.35 | 0.1 | T109, W151, N197, E198, E274, S277, R396 |
| GCP | -7.85 | 0.5 | T109, W151, N197, E198, E274, S277, R396 | |
| Zebrafish Galca | β-D-Gal | -7.14 | < 0.2 Å | G64, T109, W151, N197, E198, E274, S277 |
| GCP | -8.07 | < 0.8 Å | G64, T109, W151, N197, E198, E274, S277 | |
| Zebrafish Galcb | β-D-Gal | -7.29 | < 0.2 Å | T87, W129, N175, E176, E251, S254, R373 |
| GCP | -8.06 | < 0.8 Å | T87, W129, N175, E176, E251, S254, R373 |
Table 1. Comparison between docking and MD simulation results confirms that both β-D-Gal and GCP are stably anchored in the catalytic pocket of hGALC and the zebrafish orthologues. Docking results represent the top-ranked poses that were subsequently submitted to MD simulation. Contacts from MD simulations shown in the table are conserved in more than 90% of the simulations and in at least two out of the three replicas performed for each simulated system. Residue numbering follows the Uniprot numbering (P54803-1) for the human enzyme and the numbering for the Galca (Uniprot Q5SNX7) and Galcb (Uniprot Q7ZUD5) zebrafish orthologues.
The orientation of β-D-Gal is maintained via hydrogen bonds between the hydroxyl groups of the ligand and the catalytic residues E198 and E274 (proton donor and active site nucleophile, respectively) and the non-catalytic product binding residues T109, W151, N197, S277, and R396, with occupancy over 90% during all the replica simulations (Fig. 1A and Table 1). Simulations confirmed the ability of non-covalently bound GCP to establish stable polar contacts with all the catalytic and non-catalytic residues involved in the interaction of β-D-Gal for more than 90% of duration of the replica simulations (Fig. 1B). The orientation of GCP is further maintained by interactions with T109 and G64, which confer specificity to the ligand. The only contact not conserved upon GCP engagement is with R396 due to the lower flexibility of the epoxide compared to the hydroxyl groups exposed by the natural product at this interface (Fig. 1A,B and Table 1). This interaction is reflected in the computed binding free energy (ΔGtotal) of -46.92 ± 0.65 and − 32.05 ± 1.59 kcal/mol for β-D-Gal and GCP, respectively (Table 2). Since a detailed map of the key residues stabilizing the interaction of β-D-Gal and GCP is important for understanding its mechanism of action and for any structure-based drug design, the contribution of each residue to ΔGtotal was computed (Supplementary Fig. S1). The less favorable binding free energy of the non-covalently bound GCP compared to the natural ligand can be largely explained by the less favorable electrostatic term due to the lack of interaction with R396 (Table 2 and Supplementary Fig. S1).
Fig. 1.
Binding modes from molecular docking and molecular dynamics simulations of the interaction of β-D-Gal and GCP with human and zebrafish GALC proteins. The active site pockets of hGALC (A,B,C), zebrafish Galca (D,E,F), and zebrafish Galcb (G,H,I) proteins are shown with the orientation of β-D-Gal (left panels) and GCP (center panels) obtained after MD simulation and for GCP obtained upon covalent docking (right panels). Proteins are shown in grey cartoon representation while residues involved in the interactions are shown in stick representation colored by element with green, cyan, or turquoise carbons for hGALC, Galca, and Galcb, respectively. β-D-Gal and GCP are shown in stick representation with carbons colored brown and magenta, respectively. H-bond interactions are shown as yellow dashed lines. The covalent bond between the ligand and the protein is shown between GCP and the neutrophile residue (E274 for human GALC, E251 and E256 for zebrafish Galca and Galcb, respectively).
Table 2.
Computed binding free energy for the human and zebrafish simulated systems.
| Enzyme | Ligand | ΔGtotal | ΔGel | ΔGVdW | ΔGsolv |
|---|---|---|---|---|---|
| hGALC | β-D-Gal | -46.92 ± 0.65 | -79.58 ± 0.72 | -22.20 ± 0.76 | 54.87 ± 0.47 |
| GCP | -32.05 ± 1.59 | -48.32 ± 3.65 | -20.70 ± 3.52 | 38.51 ± 2.01 | |
| Zebrafish Galca | β-D-Gal | -43.14 ± 4.28 | -85.00 ± 3.04 | -18.91 ± 3.46 | 60.77 ± 4.63 |
| GCP | -35.36 ± 4.46 | -69.61 ± 3.83 | -19.31 ± 3.35 | 53.57 ± 4.66 | |
| Zebrafish Galcb | β-D-Gal | -42.11 ± 3.41 | -79.34 ± 7.08 | -18.41 ± 0.52 | 55.64 ± 8.67 |
| GCP | -27.71 ± 2.19 | -62.99 ± 4.12 | -17.50 ± 1.3 | 52.79 ± 2.51 |
Table 2. Molecular mechanics-generalized Born surface area (MM/GBSA) energies (kcal/mol) and their components computed for β-D-Gal or GCP bound non-covalently to hGALC, Galca, and Galcb show that the GCP inhibitor can be accommodated within the catalytic site of the enzyme with energetically favorable interactions, facilitating the formation of a covalent bond, even though these are weaker than for β-D-Gal (see Fig. 1C,F,I). The average binding free energy (ΔGtotal) is computed as the sum of the electrostatic (ΔGel), van der Waals (ΔGVdW) and solvation (ΔGsolv) -free energies. The means and standard deviations of the energies are computed from three replica simulations for each system.
Next, to obtain insights into the irreversible binding mode of GCP on GALC, the opening of the oxirane of GCP upon nucleophilic attack was investigated by performing covalent docking with Glide. The top-ranked poses show how the covalent linkage between the C3 of GCP exposed upon opening of the oxirane and the Oe2 atom of nucleophile E274 is unambiguous and only requires a minor conformational change of E274. All the interactions identified with non-covalent docking followed by MD simulations are conserved and further interactions with Y254, Y319, and R396 are established (Fig. 1C). Notably, the interaction with R396 is important for the binding of β-D-Gal (Fig. 1A). The covalent docking score is −8.72. The conservation of the contacts established with GCP prior to and upon opening of the oxirane and limited conformational changes in the active site loops indicate that the binding pocket of hGALC is highly preorganized and the flexible docking procedure followed by MD simulation enabled the identification of the orientation of GCP prior the nucleophilic attack. Indeed, upon opening of the epoxide, the RMSD computed between the GCP/GALC and the human crystal structure (PDBid 4CCE) was 0.325 Å.
Overall, our findings suggest that GCP can be accommodated in the catalytic pocket with the closed-epoxide and the subsequent nucleophilic attack by E274 triggers the formation of an irreversible linkage.
3.2. MD simulations and covalent docking elucidate the activity of the GCP inhibitor on zebrafish orthologues Galca and Galcb
The protocol described above was applied to investigate the interaction of β-D-Gal and GCP with the two GALC zebrafish orthologues Galca and Galcb [8]. The zebrafish Galca and Galcb were modelled in Swiss-Model based on the murine GALC/saposin-A crystal structure (PDBid: 5NXB) [16] and the Galca (Uniprot Q5SNX7) and Galcb (Uniprot Q7ZUD5) sequences. Flexible docking of β-D-Gal and GCP was carried out for both zebrafish orthologues simulated at pH 4.4. The top-ranked poses of β-D-Gal in Galca and Galcb overlap with those of the ligand in the murine GALC crystal structure (RMSD < 0.2 Å) while the two top-ranked poses for GCP are slightly rotated (RMSD < 0.8 Å) although they maintain the interaction with all the key residues within the catalytic pocket. Indeed, β-D-Gal interacts with T87, W129, N175, E176, E251, S254, and R373 residues in Galca and with T91, W133, N179, E180, E256, S259, and R378 residues in Galcb, while the non-covalently bound GCP establishes contacts with G42, W129, N175, E176, E251, S254, and R373 in Galca and with residues G46, W133, N179, E180, E256, S259, and R378 in Galcb (Table 1). As observed for hGALC, docking scores show better binding affinity of GCP for Galca and Galcb (Gscore= −8.07 and −8.06, respectively) when compared to the natural ligand β-D-Gal (Gscore= −7.14 and −7.29).
Multiple replica MD simulations were then performed to evaluate the stability of the systems and to confirm the binding pose of the ligands. In agreement with the docking scores, the two ligands in both zebrafish orthologues were stable along the simulations with a deviation from the starting structure of less than 0.5 Å (Fig. 1D,E,G,H and Supplementary Fig. S2). The orientation of β-D-Gal was maintained via hydrogen bonds between the hydroxyl groups of the ligand and Galca or Galcb non catalytic residues (T87, W129, N175, S254, and R373 for Galca; T91, W133, N179, S259, and R378 for Galcb) and with the proton donor and the active site nucleophile (E176 and E251 in Galca; E180 and E256 in Galcb) for more than 90% of the duration of all the replica simulations (Fig. 1D,G and Table 1). The computed binding free energy values were equal to − 43.14 ± 4.28 and − 42.11 ± 3.41 kcal/mol for β-D-Gal/Galca and β-D-Gal/Galcb, respectively (Table 2). As observed for the human model, the non-covalently bound GCP demonstrates the ability to sequester in the catalytic pocket of both zebrafish GALC orthologues, maintaining interactions with key residues for more than 90% of the simulations (Fig. 1D,E and Table 1). In the system with Galca, the orientation of GCP is stabilized by an intricate hydrogen bond network involving the hydroxyl groups of the inhibitor and T87, N175, E176, E251, and R373 residues, plus a pi-pi interaction with W284 (Fig. 1E and Table 1). In the system with Galcb, the orientation of GCP is slightly shifted and the hydrogen bond network involves the hydroxyl groups of the inhibitor and T91, Y236, E256, and R378 residues, plus a pi-pi interaction with W289 (Fig. 1H and Table 1). Remarkably, the salt bridge between the hydroxyl group of the inhibitor and the Galca residue R373 results in a more favorable electrostatic component with an increased binding free energy when compared to the GCP/hGALC complex, with a value of − 35.36 ± 4.46 kcal/mol (Table 2). Similarly, although the salt bridge involving R378 is maintained in the GCP/Galcb complex, the loss of other interactions results in a less favorable binding free energy of − 27.71 ± 2.19 kcal/mol (Table 2). Nevertheless, the less favorable binding free energy of the non-covalently bound GCP can be attributed to the less favorable electrostatic term derived from the interaction with R373 in Galca and R378 in Galcb compared to β-D-Gal (Supplementary Fig. S1).
Then, covalent docking was performed to investigate the putative formation of an irreversible linkage between GPC and Galca or Galcb upon nucleophilic attack in the catalytic pocket, as seen in hGALC. Top-ranked poses show that, upon nucleophilic attack and oxirane-opening, C4 or C3 of GPC established a covalent linkage with the Oe2 atom of E251in Galca or E256 in Galcb, respectively. In Galca, the orientation of GPC is slightly rotated compared to β-D-Gal, although the inhibitor interacts with all the pharmacophoric residues previously identified and, in addition, with G42 and Y231 (Fig. 1D,F). In Galcb, the orientation of GPC perfectly overlaps with the binding mode of β-D-Gal, and the irreversible inhibitor interacts with all the pharmacophoric residues of Galcb plus G46 (Fig. 1G,I). Covalent docking scores are − 7.37 and − 8.63 for Galca and Galcb, respectively. Supposedly, despite the preservation of the catalytic pocket in both Galca and Galcb, differences in their surrounding regions, combined with the rigidity of GCP before the oxirane opening, hindered the precise determination of the GCP orientation. However, the covalent docking results allow the identification of the interactions established by GCP on Galca and Galcb, suggesting the ability of the inhibitor to establish a covalent binding with the zebrafish orthologues in an arrangement corresponding to that seen for hGALC.
3.3. GCP inhibits GALC activity in zebrafish embryos
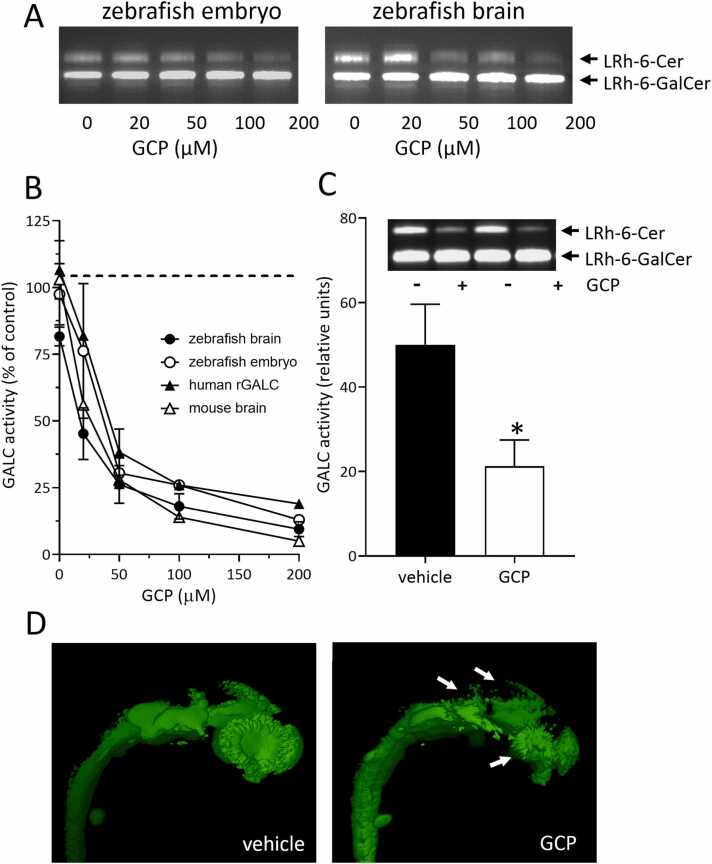
The in-silico data prompted us to assess the capacity of GCP to inhibit GALC activity in zebrafish. In a first set of experiments, the extracts of zebrafish embryos and of adult zebrafish brain were incubated with increasing concentrations of GCP. Next, GALC activity was measured by an enzymatic TLC assay using the fluorescent GALC substrate LRh-6-GalCer [28]. As shown in Fig. 2A,B, GCP inhibits the activity of GALC in zebrafish extracts in a dose-dependent manner, with a potency like that exerted on a murine brain extract or on the activity of recombinant human GALC expressed by human HEK293 cells.
Fig. 2.
Effect of GCP on GALC activity in zebrafish. A) Extracts of 96 hpf zebrafish embryos (50 μg/sample) and of adult zebrafish brain (20 μg/sample) were incubated at room temperature with increasing concentrations of GCP in the presence of the GALC substrate LRh-6-GalcCer. After overnight incubation, the GALC product LRh-6-Cer was separated from its substrate by TLC, visualized under an ultraviolet lamp, and photographed. B) Extracts of zebrafish embryos, zebrafish, and murine adult brains, and recombinant human GALC expressed in HEK293 cells (rGALC) were incubated with GCP as in A. The GALC product LRh-6-Cer was quantified following its extraction from the silica gel plates. Data are the mean ± S.E.M of 3–4 independent experiments. C) In two independent experiments, zebrafish embryos at 1–2 cell stage were injected with 160 pmoles of GCP/embryo or vehicle. At 96 hpf, 8 GCP-treated and 8 vehicle-treaded animals were pooled and processed for GALC activity assay as in panel B. Inset: Image of the TLC analysis performed on the pools of vehicle (-) or GCP (+) treated animals. P, paired Student’s t test. D) Light sheet 3D reconstruction of Tg(neurod1:EGFP)ia50 embryos injected at 6 hpf with 160 pmoles/embryo of GCP or vehicle and visualized at 24 hpf. After image acquisition, 3D reconstructions were performed using the Arivis software (Zeiss) and exported as a single snap with the same compression settings. White arrows indicate the midbrain and eye regions of the embryos.
On this basis, the impact of GCP on GALC activity was assessed in vivo in zebrafish embryos. To this aim, zebrafish embryos at 1–2 cell stage were microinjected with vehicle (10% DMSO in water) or with the maximal tolerated dose of GCP dissolved in vehicle (equal to 160 pmoles/embryo). After 96 h, no significant differences in embryo mortality and macroscopic morphologic alterations were observed between the two experimental groups (data not shown) and GALC activity was assessed in the embryo extracts by the enzymatic TLC assay. In keeping with its irreversible mechanism of action, a significant decrease in GALC activity was observed in GCP-treated embryos when compared to controls (Fig. 2C). Similar results were obtained when GCP was injected in zebrafish embryos at shield stage (data not shown).
Double galca/galcb zebrafish embryo morphants are characterized by the partial disorganization of the expression of the neuronal marker neurod1 in the central nervous system because of the simultaneous downregulation of both galc orthologues [8]. In keeping with these observations, GCP administration in Tg(neurod1:EGFP)ia50 embryos [26] causes a significant alteration of the spatial neurod1 expression in the midbrain and eye regions as assessed by 3D light sheet microscopy (Fig. 2D).
3.4. GCP modulates the lipid profile of zebrafish embryos
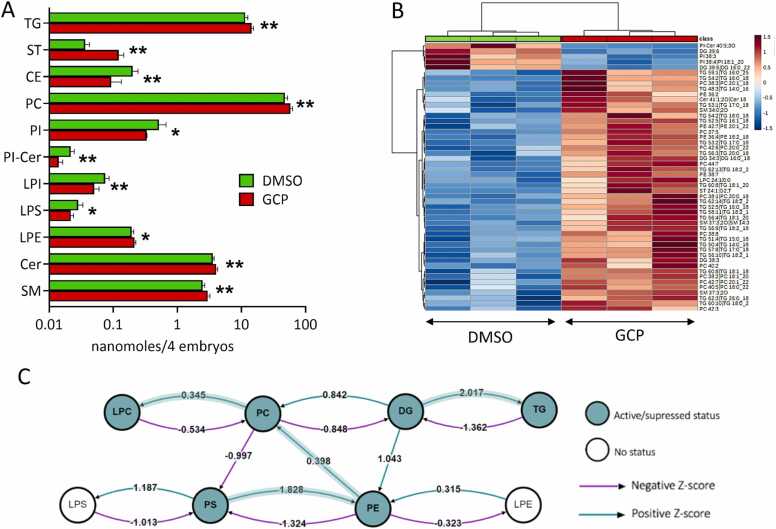
Untargeted lipidomic analysis was used to assess the effect of GALC inhibition on the lipid composition in 96 hpf embryos microinjected at 1–2 cell stage with 160 pmoles/embryo of GCP or vehicle. The analysis identified 766 lipid species in both experimental groups, representing various classes of lipids that included: bis(monoacylglycerol)phosphate (BMP, 1 species), fatty acids (FA, 8 species), fatty acyl carnitines (CAR, 17 species), N-acyl-ethanolamines (NAE, 9 species), sterols (ST, 3 species), free cholesterol, cholesteryl esters (CE, 2 species), diacylglycerols (DG, 58 species), and triglycerides (TG, 179 species); the sphingolipids ceramides (Cer, 20 species), sphingomyelins (SM, 52 species), and sphingoid bases (SPB, 1 species); the phospholipids phosphatidylcholines (PC, 193 species), phosphatidylethanolamines (PE, 102 species), phosphatidylglycerols (PG, 2 species), phosphatidylinositols (PI, 39 species), and phosphatidylserines (PS, 24 species); the lysophospholipids lyso-PC (LPC, 27 species), lyso-PE (LPE, 15 species), lyso-PI (LPI, 5 species), and lyso-PS (LPS, 3 species) (Supplementary Table S1).
When GCP-treated embryos were compared to vehicle-treated animals, significant differences were observed for the levels of TG and ST classes, as well as for the levels of Cer and SM, all increased in GCP-treated animals. By contrast, the levels of CE were decreased by GCP. In addition, significant changes were observed for the levels of various classes of phospholipids. They included an increase in the amount PC and LPE, paralleled by a decrease in PI, LPI, LPS, and PI-Cer levels (Fig. 3A). Moreover, analysis of the metabolic pathways affected by GCP treatment using the BioPan platform [31] identified 77 activated and 43 suppressed reactions. Among them, DG→TG and PS→PE→PC→LPC pathways accounted for 68% of the activated reactions (53/77) whereas TG→DG and PE→PS represented 50% of the suppressed reactions (21/43) (Fig. 3C and Supplementary Table S2).
Fig. 3.
Effect of GCP on the lipid profile of zebrafish embryos. A) Levels of the different classes of lipids detected in DMSO and GCP-treated embryos. Embryos were treated at the 1–2 cell stage and lipid analysis was performed at 96 hpf. Data are expressed as nanomoles/pool of 4 embryos. * , P < 0.1; * *, P < 0.05, Student’s t test. B) Heatmap showing hierarchical clustering of the lipid species between GCP-treated zebrafish embryos and control animals (DMSO). Only the 50 most important lipids species are displayed based on their t-test p-values. Color coding represents the -Log p-value. C) Lipid subclass correlation network of GCP-treated embryos compared to controls.
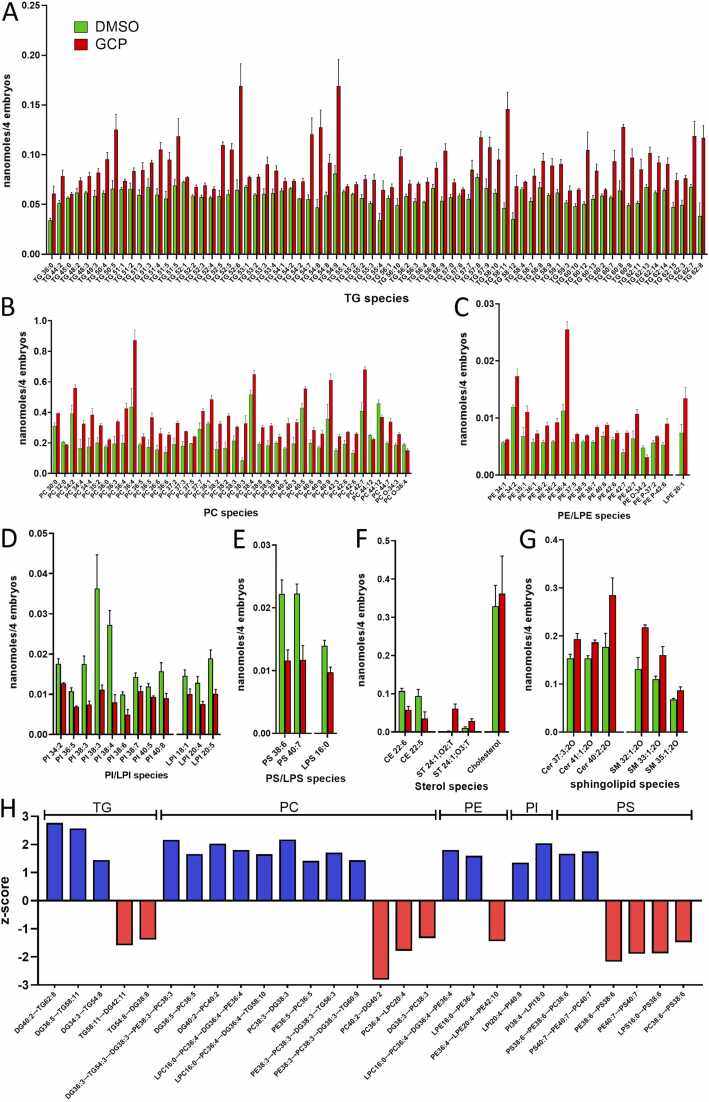
Analysis of the individual molecular species confirmed the capacity of GCP to modulate the lipid composition of zebrafish embryos. Indeed, among the 766 lipid species identified in the two experimental groups, 186 lipids were present at different levels in GCP-treated embryos when compared to controls (Fig. 3B and Supplementary Table S3). Many of these lipid species belong to the TG class [74/179 (41%)] and to the PC class [42/193 (22%)] (Fig. 4A,B). Among them, BioPan analysis pointed to the activation of the synthesis of TG 62:8, 58:11, and 54:8 species from DG 40:2, 36:5, and 34:3 species, respectively, whereas the degradation of TG 58:11 and 54:8 to DG 42:11 and 38:8 was suppressed. Similarly, the production of PC 40:2, 38:3, 36:5 and 36:4 species was increased whereas the degradation of PC 40:2 and 36:4 to DG 40:2 and LPC 20:4 was suppressed (Fig. 4H).
Fig. 4.
GCP affects the levels of various lipid species in zebrafish embryos. A-G) Levels of the different species of lipids whose amount was significantly affected by GCP treatment when compared to controls. Embryos were treated at the 1–2 cell stage and lipid analysis was performed at 96 hpf. Data are expressed as nanomoles/pool of 4 embryos. H) Z-score representation of the reactions that involve the most GCP-modulated species of lipids following BioPan analysis of lipidomic data. Positive z-score indicates an activated reaction, negative z-score indicates a suppressed reaction.
Lipid species in classes other than TG and PC were affected as well. Among phospholipid classes other than PC, various PE species (17/102; 16%) were significantly increased, including PE 36:4 (more than two-fold increase), and PE 42:7 plus the corresponding LPE 20:1 (both increased by 80%). Accordingly, PE 36:4 conversion from LPC 16:0 was activated in parallel with the suppression of its degradation to LPE 20:4 (Fig. 4C,H). Conversely, 9 out of 39 PI species (23%) and 3 out of 5 LPI species (60%) were decreased following GCP administration. Among them, the PI 38:4 and 38:3 species were decreased by 70% because of an augmented conversion to LPI 18:0 whereas LPI 20:5 and 20:4 species decreased by approximately 50%, LPI 20:4 being converted to PI 40:8 (Fig. 4D,H). Similarly, PS 38:6 and 40:7, as well as LPS 16:0, were also decreased by 30 to 50% following their conversion to PC 38:6 and 40:7, respectively, together with a reduction in their synthesis reactions (Fig. 4E,H). As for sterols, CE 22:5 and 22:6 were decreased by 40% and 50% respectively, whereas ST 24:1;O2;T and 24:1;O3;T were increased significantly, no changes being observed for free cholesterol (Fig. 4F). Finally, among sphingolipids, the levels of 3 out of 20 Cer species were more abundant in GCP-treated embryos, Cer 40:2;20, 37:3;20, and 41:1;20 species being increased by more than 60%, 25%, and 20%, respectively. Similarly, among 52 SM species, the levels of SM 32:1;20, 33:1;20, and 35:1;20 were increased by more than 50% following GCP treatment (Fig. 4G). Together, the data indicate that inhibition of GALC activity by GCP exerts a significant impact on the lipid profile of zebrafish embryo.
4. Discussion
The β-galactopyranose-configured cyclophellitol-epoxide GCP has been demonstrated to inhibit the enzymatic activity of recombinant and rodent GALC covalently and irreversibly [11]. Here, docking studies followed by MD simulations indicate that GCP can bind in the catalytic pocket of hGALC and of the zebrafish orthologues Galca and Galcb with the closed epoxide (pre-reactive GCP). Within the catalytic pocket, GCP shares the same pharmacophoric interaction points as the natural product β-D-Gal, indicating its capacity to exert a competitive inhibitory effect. Contact analysis and per-residue energy decomposition studies unveiled a greater abundance of polar contacts contributing to the binding of the natural substrate β-D-Gal compared to the non-covalently bound inhibitor GCP. This translates into a substantial difference in the binding free energy between the compounds, as obtained by the MM/GBSA method, with more favorable binding free energy for the natural ligand than for the inhibitor in all the systems studied. Notably, although the MM/GBSA method offers valuable insights into molecular interactions, the ΔGtotal values obtained may be overestimated due to inherent limitations of the methodology [24], [32]. Nevertheless, MM/GBSA is a commonly used approach, and its results are often consistent with trends in measured Kd values, providing relative rather than absolute values for comparison of binding free energy between ligands [24], [32]. Finally, the contrasting trend observed between the Glide docking and MMGBSA results can be attributed to the different scoring functions utilized in the two methods: the former rewards non-polar interactions - favorable for the GCP inhibitor - while the latter prioritizes more polar interactions, favoring the natural ligand. Since a detailed description of the irreversible binding of GCP is important for understanding its mechanism of action, covalent docking was also performed. These dockings reveal that the lysosomal acid environment promotes the protonation of the oxirane while the nucleophile residue within the catalytic pocket (E274, E251, and E256 in hGALC, Galca, and Galcb, respectively) triggers its opening and the formation of a covalent bond, thus leading to irreversible inhibition of human and zebrafish enzymes. Remarkably, despite the small differences in the catalytic pocket of the human and zebrafish orthologues, GCP is predisposed to form a covalent bond in the case of the human GALC as well as for the zebrafish orthologues. Overall, based on our in-silico predictions, we demonstrated that once the pre-reactive GCP closed-epoxide is accommodated within the catalytic site of the enzyme, the close proximity between the reactive epoxide of the inhibitor and the side chain of the nucleophile residue triggers the formation of a covalent bond, making the GCP an irreversible inhibitor. The less favorable MM/GBSA free energies of the pre-reactive GCP compared to the natural ligand indicate that the covalent bond formation is necessary to exert an inhibitory effect.
In keeping with this hypothesis, GCP inhibits the enzymatic activity of GALC present in zebrafish embryos and adult brain extracts with a potency corresponding to that exerted on human and murine GALC. Accordingly, injection of GCP into zebrafish embryos at 1–2 cell stage leads to a significant decrease in GALC activity that was retained for at least 96 h after injection. On this basis, we analyzed the impact of GALC inhibition on the lipid profile of zebrafish embryos.
Untargeted lipidomic analysis identified 766 lipid species in the whole extracts of GCP-treated and control zebrafish embryos at 96 hpf. At this developmental stage, the lipid content of the yolk sac is remarkably decreased when compared to the embryo body [33]. Thus, the results of our analysis reflect the effect of GCP on the lipid metabolism in the embryo proper, with a minor contribution due to alterations of lipid remodeling in the yolk sac. This might be the case for the observed increase in the levels of 74 out of 179 TG species following GCP treatment, consequent to a reduced use of the yolk TG by the embryo body.
In keeping with the decrease in CE levels observed in murine melanoma B16-F10 cells after Galc knockdown [34], GCP treatment caused a decrease in the amount of the CE species 22:5 and 22:6 in zebrafish embryos with a consequent decrease in the total levels of CE. CE reduction in GCP-treated embryos was paralleled by an increase in the levels of the taurine-conjugated bile acids ST 24:1;O2;T and 24:1;O3;T, with no changes in free cholesterol. Maturation of the liver occurs in zebrafish embryos at 72–120 hpf [35]. In keeping with the alterations observed in the liver of adult GALC-deficient twitcher mice, an authentic model of Krabbe disease [36], [37], these data suggest that a decrease in GALC activity may affect liver metabolism in zebrafish embryos.
Notably, significant changes were observed for the levels of various classes of phospholipids following GCP treatment. They included an increase in PC, PE, and LPE levels, paralleled by a decrease of PI, LPI, LPS, and PI-Cer. As observed for TG changes, the effect of GCP on these phospholipid classes was restricted to defined lipid species, pointing again to a specific effect of GALC inhibition on phospholipid metabolism in zebrafish embryos. In keeping with these observations, a significant reduction of PE was observed in Galc knockdown B16 cells [34] and alterations of the phospholipid profile and membrane turnover have been reported in the brain of twitcher mice [38] as a possible consequence of the tight crosstalk that occurs between phospholipid and sphingolipid metabolism [reviewed in [39]]. Indeed, significant changes in the levels of Cer and SM were detected in GCP-treated animals. They included Cer 40:2;20, 37:3;20, and 41:1;20, as well as SM 32:1;20, 33:1;20, and 35:1;20, the increased amount of SM reflecting the higher content of its biosynthetic precursors PC and Cer.
The effect of GCP on Cer deserves further discussion. Previous observations had shown that the total levels of Cer are decreased in Galc null murine hematopoietic stem cells [40] whereas they are increased in Galc knockdown murine melanoma B16 cells [34]. On the other hand, a lipidomic approach has shown that the levels of Cer 16:0 were increased in the central nervous system of twitcher mice when compared to wildtype animals, whereas the levels of Cer 18:0, 22:0, and 24:0 were reduced [41]. The Cer species affected by GCP in zebrafish belong to both long acyl chain (C18:0) and very long acyl chain (22:0; 23:0, and 24:0) species. Nine highly conserved homologs of the vertebrate Cer synthase gene family have been identified in the zebrafish genome [42]. Since the six members of the mammalian Cer synthase family show substrate preference for FA chains of different length [43], it seems possible to hypothesize that GALC deficiency may affect different Cer synthases in a context-dependent manner, with a consequent impact on different Cer species.
GCP is a competitive and irreversible GALC inhibitor with no anti-α-galactosidase nor anti-β-glucosidase activity, its selectivity arising from its absolute configuration [11], [44]. However, we cannot rule out the possibility that its effect on the zebrafish embryo lipidome may be due in part also on its possible interaction with β-galactosidase(s) distinct from GALC, including acid β-galactosidase (BGAL) [11]. Relevant to this point, it has been demonstrated that the crossing of GALC-deficient twitcher mice with BGAL deficient animals leads to paradoxical effects [45]. Indeed, heterozygous Bgal-/+ mice in a Galc null background show a more severe phenotype when compared to double Galc/Bgal knockout animals. Of note, Galc-/-/Bgal-/+ mice are characterized by a dramatic increase of lactosylceramide that we did not detect in GCP-treated zebrafish embryos, suggesting that the contribution of BGAL inhibition by GCP might be negligible (if any). Experiments performed on double galca/galcb null zebrafish embryos will be required to elucidate this point.
No significant alterations in embryo survival and morphology appear to occur in animals treated with the maximal tolerated dose of GCP. Nevertheless, as observed in double galca/galcb zebrafish morphants [8], GCP administration causes significant alterations of the spatial expression of the neuronal marker neurod1 in zebrafish embryos, as assessed by light sheet microscopy using the transgenic reporter Tg(neurod1:EGFP)ia50 zebrafish line. These data indicate that GALC deficiency may exert a significant impact on the development of the central nervous system also during embryonic development. The lack of GALC activity in Krabbe patients and twitcher mice is characterized by a progressive increase in the neurotoxic GALC substrate psychosine after birth. Here, the levels of hexosylsphingosines (including psychosine) and lactosylceramide were below the limits of detection in both control and GCP-treated embryos. These data are in keeping with the lack of psychosine accumulation in double galca/galcb zebrafish embryo morphants and human Krabbe fetuses [8], [9], [10], possibly because of the early developmental time at which such analyses were performed and/or because of the residual GALC activity retained after GCP or morpholino injection. Together, these observations support the hypothesis that the loss of GALC may have pathological consequences during embryonic development that are independent of psychosine accumulation.
Thus far, only limited information has been available about the effect of the modulation of GALC activity on the lipidome, mainly obtained by the analysis of tissues harvested from adult twitcher mice or genetically modified cell lines [reviewed in [46]]. In addition, even though scattered evidence indicates that alterations of the central and peripheral nervous system are observed in human Krabbe fetuses [9], [10], no data were available about the effects of the lack of GALC activity on the lipidome of developing organisms. Our data suggest that GALC may play a non-redundant role in lipid metabolism during embryonic development before the accumulation of significant levels of neurotoxic psychosine may occur. Thus, this work provides insights into the pathogenesis of Krabbe disease and the basis for a better comprehension of the alterations of the central and peripheral nervous system observed in human Krabbe fetuses.
Funding sources
This work was supported by “Associazione Italiana per la Ricerca sul Cancro” [AIRC IG 23116 to M.P] and by NextGenerationEU [PNRR M4C2-Investimento 1.4- CN00000041]. J.C. is supported by an AIRC fellowship [ID 26633]. M.M. is supported by the AGING Project – Department of Excellence – DIMET, Università del Piemonte Orientale. G.P. and R.C.W. thank the Klaus Tschira Foundation. G.P was supported by a Joachim Herz Foundation Fellowship and by the Artificial Intelligence (AI) Health Innovation Cluster.
CRediT authorship contribution statement
Mirella Belleri: Investigation. Giulia Paiardi: Formal analysis, Investigation, Methodology, Writing – original draft. Chiara Tobia: Investigation. Elisa Scalvini: Investigation. Davide Capoferri: Data curation. Marzia Corli: Investigation. Rebecca C. Wade: Supervision, Writing – review & editing. Marco Ghirimoldi: Investigation. Marcello Manfredi: Data curation. Marco Presta: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Luca Mignani: Data curation, Investigation, Writing – review & editing. Jessica Guerra: Conceptualization, Investigation, Methodology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The GALC inhibitor GCP was kindly provided by M. Artola and H.S. Overkleeft (Department of Medical Biochemistry, Leiden Institute of Chemistry, Leiden University, Leiden, The Netherlands). This work was supported by “Associazione Italiana per la Ricerca sul Cancro” [AIRC IG 23116 to M.P] and by NextGenerationEU [PNRR M4C2-Investimento 1.4- CN00000041]. J.G. is supported by an AIRC fellowship [ID 26633]. M.M. is supported by the AGING Project – Department of Excellence – DIMET, Università del Piemonte Orientale. G.P. and R.C.W. thanks the Klaus Tschira Foundation. G.P was supported by a Joachim Herz Foundation Fellowship and by the Artificial Intelligence Health Innovation Cluster.
All the experiment on Danio rerio were performed in the Zebrafish facility of the DMMT, University of Brescia, Italy.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.03.023.
Appendix A. Supplementary material
Supplementary material
.
Data availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Bradbury A.M., Bongarzone E.R., Sands M.S. Krabbe disease: new hope for an old disease. Neurosci Lett. 2021;752 doi: 10.1016/j.neulet.2021.135841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon J.M., Matern D., Kurtzberg J., Wrabetz L., Gelb M.H., Wenger D.A., et al. Consensus guidelines for newborn screening, diagnosis and treatment of infantile Krabbe disease. Orphanet J Rare Dis. 2018;13:30. doi: 10.1186/s13023-018-0766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon I.C., Bascou N.A., Poe M.D., Szabolcs P., Escolar M.L. Long-term neurodevelopmental outcomes of hematopoietic stem cell transplantation for late-infantile Krabbe disease. Blood. 2021;137:1719–1730. doi: 10.1182/blood.2020005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Xu Y., Benitez B.A., Nagree M.S., Dearborn J.T., Jiang X., et al. Genetic ablation of acid ceramidase in Krabbe disease confirms the psychosine hypothesis and identifies a new therapeutic target. Proc Natl Acad Sci USA. 2019;116:20097–20103. doi: 10.1073/pnas.1912108116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafi M.A. Krabbe disease: a personal perspective and hypothesis. Bioimpacts. 2022;12:3–7. doi: 10.34172/bi.2021.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki K., Suzuki K. The twitcher mouse: a model for Krabbe disease and for experimental therapies. Brain Pathol. 1995;5:249–258. doi: 10.1111/j.1750-3639.1995.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 7.Mignani L., Guerra J., Corli M., Capoferri D., Presta M. Zebra-Sphinx: modeling sphingolipidoses in zebrafish. Int J Mol Sci. 2023;24:4747. doi: 10.3390/ijms24054747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zizioli D., Guarienti M., Tobia C., Gariano G., Borsani G., Bresciani R., et al. Molecular cloning and knockdown of galactocerebrosidase in zebrafish: new insights into the pathogenesis of Krabbe's disease. Biochim Biophys Acta. 2014;1842:665–675. doi: 10.1016/j.bbadis.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Suchlandt G., Schlote W., Harzer K. Ultrastructural findings in 9 fetuses following prenatal diagnosis of neurolipidoses. Arch Psychiatr Nervenkr (1970) 1982;232:407–426. doi: 10.1007/bf00345597. [DOI] [PubMed] [Google Scholar]
- 10.Martin J.J., Leroy J.G., Ceuterick C., Libert J., Dodinval P., Martin L. Fetal Krabbe leukodystrophy. A morphologic study of two cases. Acta Neuropathol. 1981;53:87–91. doi: 10.1007/bf00689987. [DOI] [PubMed] [Google Scholar]
- 11.Marques A.R., Willems L.I., Herrera Moro D., Florea B.I., Scheij S., Ottenhoff R., et al. A specific activity-based probe to monitor family GH59 galactosylceramidase, the enzyme deficient in krabbe disease. Chembiochem. 2017;18:402–412. doi: 10.1002/cbic.201600561. [DOI] [PubMed] [Google Scholar]
- 12.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. doi: 10.1093/nar/gky427%J Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill C.H., Graham S.C., Read R.J., Deane J.E. Structural snapshots illustrate the catalytic cycle of β-galactocerebrosidase, the defective enzyme in Krabbe disease. Proc Natl Acad Sci USA. 2013;110:20479–20484. doi: 10.1073/pnas.1311990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill C.H., Cook G.M., Spratley S.J., Fawke S., Graham S.C., Deane J.E. The mechanism of glycosphingolipid degradation revealed by a GALC-SapA complex structure. Nat Commun. 2018;9(1):151. doi: 10.1038/s41467-017-02361-y. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 17.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pKaprediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 18.Case, D.A., Ben-Shalom, I.Y., Brozell, S.R., Cerutti, D.S., Cheatham, T.E.I.I.I., Cruzeiro, V.W.D., et al., 2018. AMBER 18. San Francisco, CA, USA: University of California.
- 19.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 21.Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 23.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 3rd ed., University of Oregon Press,; Eugene: 1995. [Google Scholar]
- 26.Mueller T., Wullimann M.F. Expression domains of neuroD (nrd) in the early postembryonic zebrafish brain. Brain Res Bull. 2002;57(3-4):377–379. doi: 10.1016/s0361-9230(01)00694-3. [DOI] [PubMed] [Google Scholar]
- 27.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 28.Marchesini S., Preti A., Aleo M.F., Casella A., Dagan A., Gatt S. Synthesis, spectral properties and enzymatic hydrolysis of fluorescent derivatives of cerebroside sulfate containing long-wavelength-emission probes. Chem Phys Lipids. 1990;53:165–175. doi: 10.3390/ijms241310555. [DOI] [PubMed] [Google Scholar]
- 29.Consonni F.M., Durante B., Manfredi M., Bleve A., Pandolfo C., Garlatti V., et al. Immunometabolic interference between cancer and COVID-19. Front Immunol. 2023;14 doi: 10.3389/fimmu.2023.1168455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng J., Shirihai O.S., Grinstaff M.W. Modulating lysosomal pH: a molecular and nanoscale materials design perspective. J Life Sci. 2020;2:25–37. doi: 10.36069/jols/20201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaud C., B C.S., Nguyen A., Fedorova M., Ni Z., O'Donnell V.B., et al. BioPAN: a web-based tool to explore mammalian lipidome metabolic pathways on LIPID MAPS. F1000Res. 2021;10:4. doi: 10.12688/f1000research.28022.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 33.Fraher D., Sanigorski A., Mellett N.A., Meikle P.J., Sinclair A.J., Gibert Y. Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Rep. 2016;14:1317–1329. doi: 10.1016/j.celrep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Belleri M., Paganini G., Coltrini D., Ronca R., Zizioli D., Corsini M., et al. β-galactosylceramidase promotes melanoma growth via modulation of ceramide metabolism. Cancer Res. 2020;80:5011–5023. doi: 10.1158/0008-5472.CAN-19-3382. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Y.C., Wu T.S., Huang Y.T., Chang Y., Yang J.J., Yu F.Y., et al. Aflatoxin B1 interferes with embryonic liver development: involvement of p53 signaling and apoptosis in zebrafish. Toxicol. 2021;458 doi: 10.1016/j.tox.2021.152844. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T., Yamanaka T., Jacobs J.M., Teixeira F., Suzuki K. The twitcher mouse: an enzymatically authentic model of human globoid cell leukodystrophy (Krabbe disease) Brain Res. 1980;202:479–483. doi: 10.1016/0006-8993(80)90159-6. [DOI] [PubMed] [Google Scholar]
- 37.Contreras M.A., Haq E., Uto T., Singh I., Singh A.K. Psychosine-induced alterations in peroxisomes of twitcher mouse liver. Arch Biochem Biophys. 2008;477:211–218. doi: 10.1016/j.abb.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinstock N.I., Wrabetz L., Feltri M.L., Shin D. Metabolic profiling reveals biochemical pathways and potential biomarkers associated with the pathogenesis of Krabbe disease. J Neurosci Res. 2016;94:1094–1107. doi: 10.1002/jnr.23789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Cuenca S., Pellegrinelli V., Campbell M., Oresic M., Vidal-Puig A. Sphingolipids and glycerophospholipids - the "ying and yang" of lipotoxicity in metabolic diseases. Prog Lipid Res. 2017;66:14–29. doi: 10.1016/j.plipres.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Visigalli I., Ungari S., Martino S., Park H., Cesani M., Gentner B., et al. The galactocerebrosidase enzyme contributes to the maintenance of a functional hematopoietic stem cell niche. Blood. 2010;116:1857–1866. doi: 10.1182/blood-2009-12-256461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esch S.W., Williams T.D., Biswas S., Chakrabarty A., LeVine S.M. Sphingolipid profile in the CNS of the twitcher (globoid cell leukodystrophy) mouse: a lipidomics approach. Cell Mol Biol. 2003;49:779–787. [PubMed] [Google Scholar]
- 42.Brondolin M., Berger S., Reinke M., Tanaka H., Ohshima T., Fuβ B., et al. Identification and expression analysis of the zebrafish homologs of the ceramide synthase gene family. Dev Dyn. 2013;242:189–200. doi: 10.1002/dvdy.23913. [DOI] [PubMed] [Google Scholar]
- 43.Ho Q.W.C., Zheng X., Ali Y. Ceramide acyl chain length and its relevance to intracellular lipid regulation. Int J Mol Sci. 2022;23 doi: 10.3390/ijms23179697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schröder S.P., van de Sande J.W., Kallemeijn W.W., Kuo C.L., Artola M., van Rooden E.J., et al. Towards broad spectrum activity-based glycosidase probes: synthesis and evaluation of deoxygenated cyclophellitol aziridines. Chem Commun. 2017;53:12528–12531. doi: 10.1039/c7cc07730k. [DOI] [PubMed] [Google Scholar]
- 45.Tohyama J., Vanier A.T., Suzuki K., Ezoe T., Matsuda J., Suzuki K. Paradoxical influence of acid β-galactosidase gene dosage on phenotype of the twitcher mouse (genetic galactosylceramidase deficiency) Hum Mol Genet. 2000;9:1699–1707. doi: 10.1093/hmg/9.11.1699. [DOI] [PubMed] [Google Scholar]
- 46.Belleri M., Chiodelli P., Corli M., Capra M., Presta M. Oncosuppressive and oncogenic activity of the sphingolipid-metabolizing enzyme β-galactosylceramidase. Biochim Biophys Acta Rev Cancer. 2022;1877 doi: 10.1016/j.bbcan.2021.188675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.