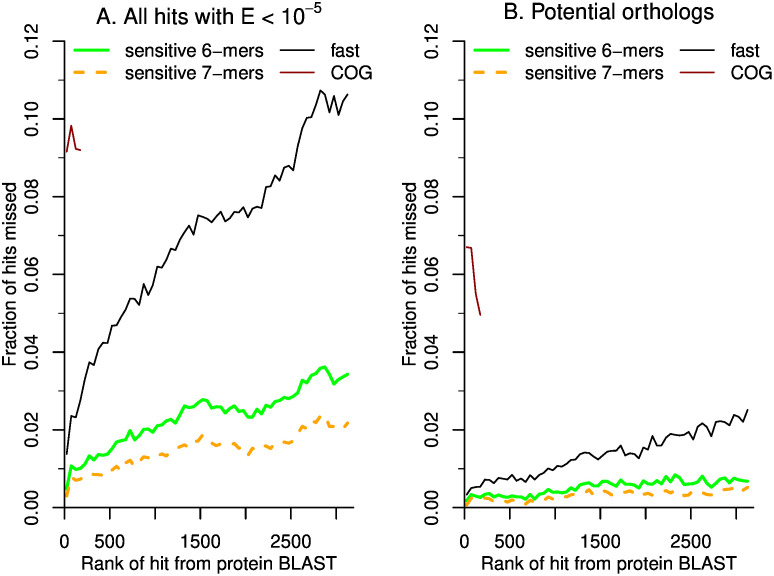
Fig 1. Sensitivity of fast parallel MMseqs2 when searching against 6,377 representative genomes.
(A) The miss rate as a function of the hit’s rank, for hits from BLAST+ with E ≤ 10−5. Each point is an average across 50 ranks and up to 1,000 queries. Besides fast parallel MMseqs2, we also show results for MMseqs2 with the most sensitive settings (using either 6-mers or 7-mers) and for COGs (as assigned using eggNOG-mapper). For COGs, we only considered the top 200 hits of each query. (B) The miss rate for potential orthologs (at least 30% identity and at least 50% coverage of the query).