Extended Data Figure 8. IRF1 and ZNF217 CUT&RUN, ChIP-Seq, and Vγ9Vδ2 T cell killing.
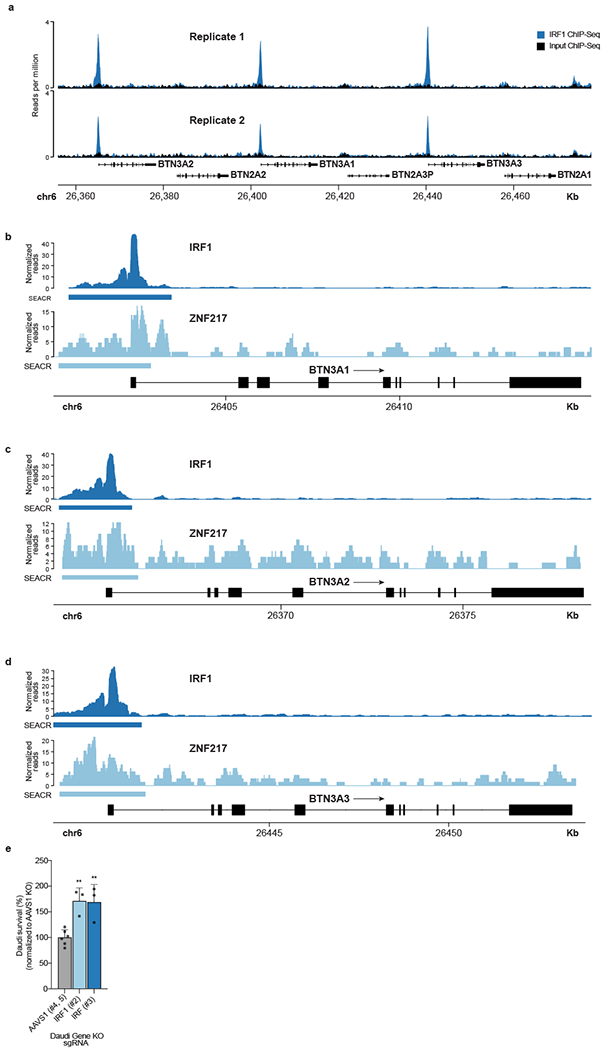
(a) Publicly available IRF1 ChIP-Seq for the butyrophilin locus in K562 cells stably expressing C-terminal eGFP-tagged IRF1 (ENCODE). (b-d) CUT&RUN data for IRF1 and ZNF217 binding at promoters in (b) BTN3A1, (c) BTN3A2, and (d) BTN3A3 loci in WT Daudi-Cas9 cells. n=3 per condition. The algorithm SEACR calls peaks and verifies them above a stringent background signal threshold. (e) Daudi-Cas9 KO survival after 24-hour co-culture with expanded Vγ9Vδ2 T cells in the presence of ZOL at an E:T ratio of 2:1. For each γδ T cell donor, Daudi survival is calculated relative to Daudi cells cultured without T cells and normalized to the Daudi-Cas9 AAVS1 KO control cell survival. Combined data from three donors and two independent experiments. AAVS1 KO n=6, IRF1 KO n=3. One-way ANOVA comparison to AAVS1 KO cells with Dunnett’s multiple comparisons test. Mean ± SD. p<0.01 (**).
