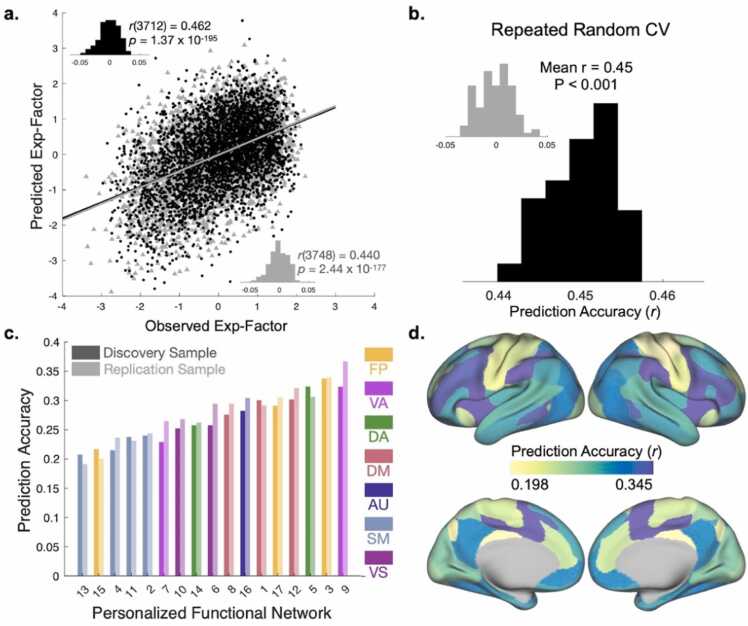
Fig. 3.
Exposome scores are reflected in the multivariate pattern of personalized functional brain network topography. (A) Association between observed and predicted general exposome factor scores using two-fold cross-validation (2 F-CV) across both the discovery (black scatterplot) and replication (gray scatterplot) samples. Inset histograms represent null distributions of prediction accuracies with permuted data. (B) Repeated random 2 F-CV provided evidence of stable prediction accuracy across splits of the data, compared with a null distribution with permuted data (inset). (C) Independent network models ranked by prediction accuracy. Note that all p-values associated with prediction accuracies are significant after Bonferroni correction. Numerical assignments for each PFN are consistent with previous work (Cui et al., 2020, Keller et al., 2023a) and can be found in Fig. S4. (D) Prediction accuracy averaged across discovery and replication samples depicted for seventeen cross-validated models trained on each PFN independently. Abbreviations: Exp-Factor: general exposome factor; CV: cross-validation; FP: fronto-parietal; VA: ventral attention; DA: dorsal attention; DM: default mode; AU: auditory; SM: somatomotor; VS: visual.