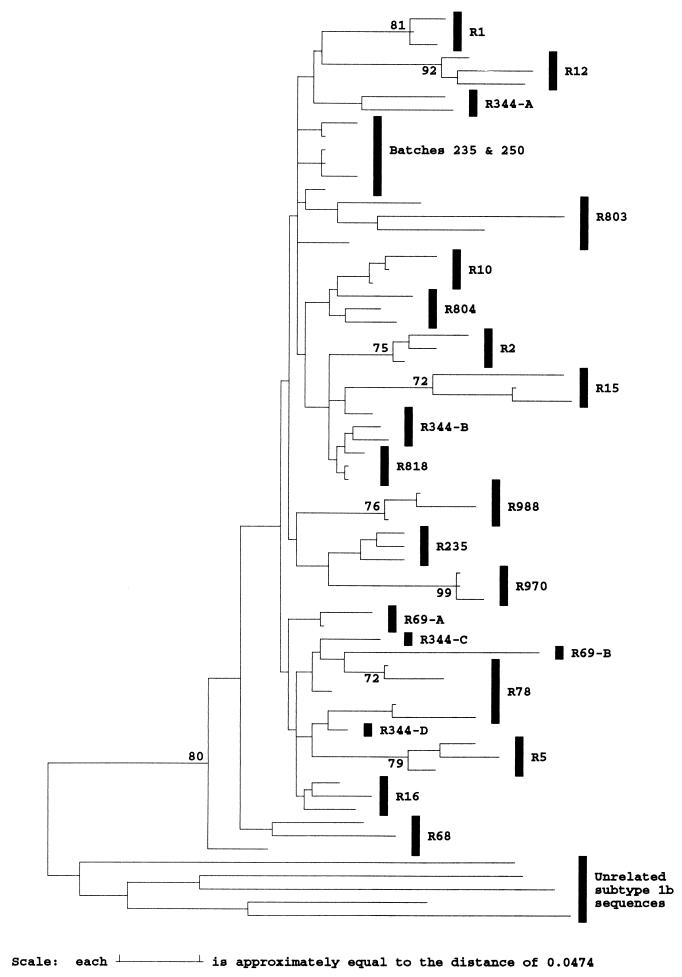
FIG. 5.
Phylogenetic analysis of the HVR flanking region at synonymous sites. Evolutionary distances between sequences at synonymous sites between positions 1096 to 1150 and 1231 to 1458 were calculated with the Jukes-Cantor correction for representative sequences from different anti-D recipients (R), two infectious batches (238 and 250), and five unrelated subtype 1b sequences and used to construct a neighbor-joining tree. Sequences from the same recipient are grouped by vertical bars, except where these group separately on the tree, in which case a suffix indicates the HVR group to which the sequences belong. Bootstrap values of 70% or more (500 replicates) are indicated.