Fig. 1. Brain tissue samples used for data collection and initial clustering of snRNA-seq data.
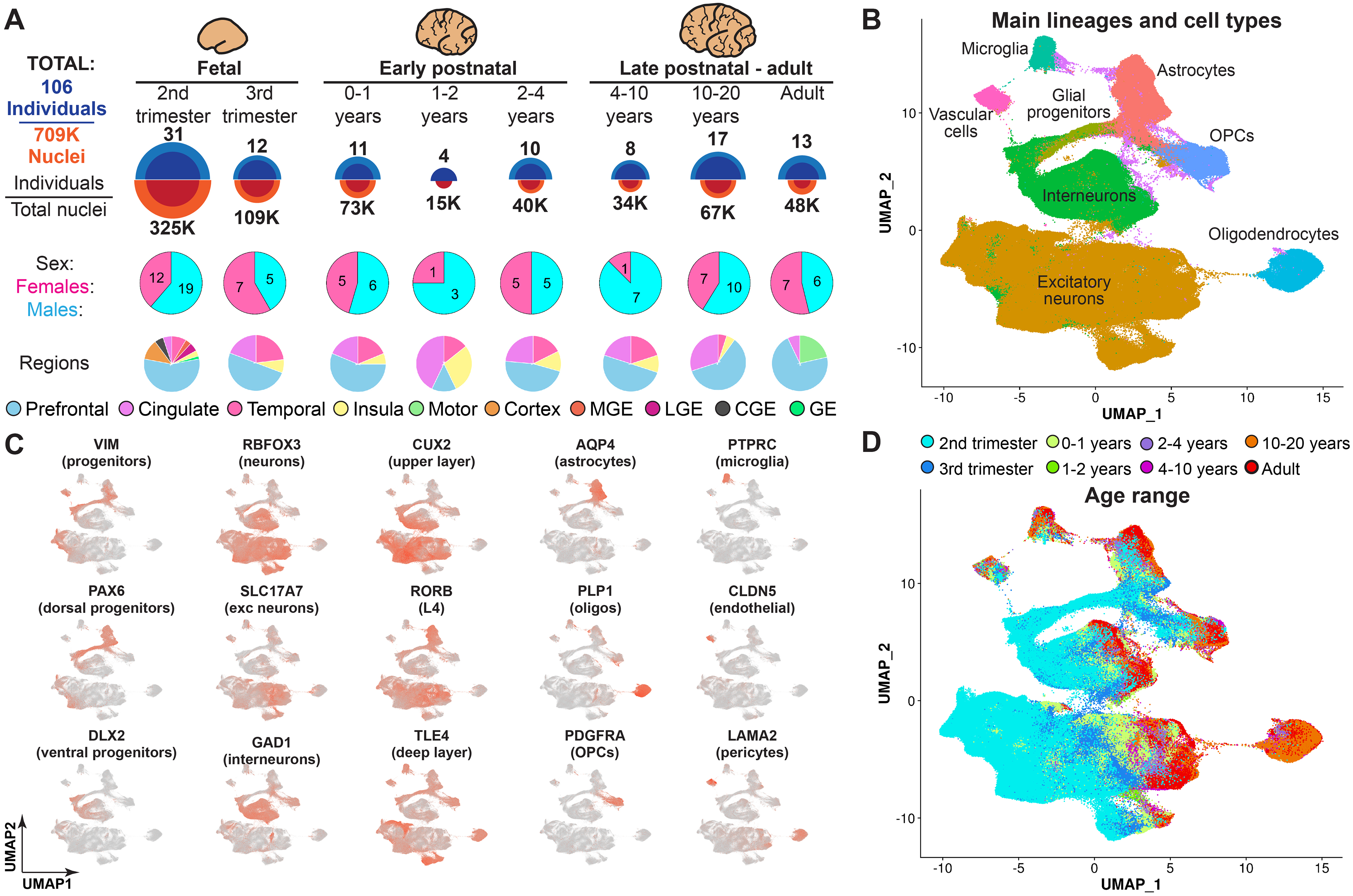
(A) Overview of the tissue samples used in this study, including the number of individuals and the ages and brain regions captured in the snRNA-seq dataset. MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence; CGE, caudal ganglionic eminence; GE, ganglionic eminence. (B) Clustering of the entire dataset, with the major lineages labeled. (C) Expression of cell type–specific markers used to determine cardinal lineages. exc neurons, excitatory neurons. (D) Nuclei labeled by their developmental age.
