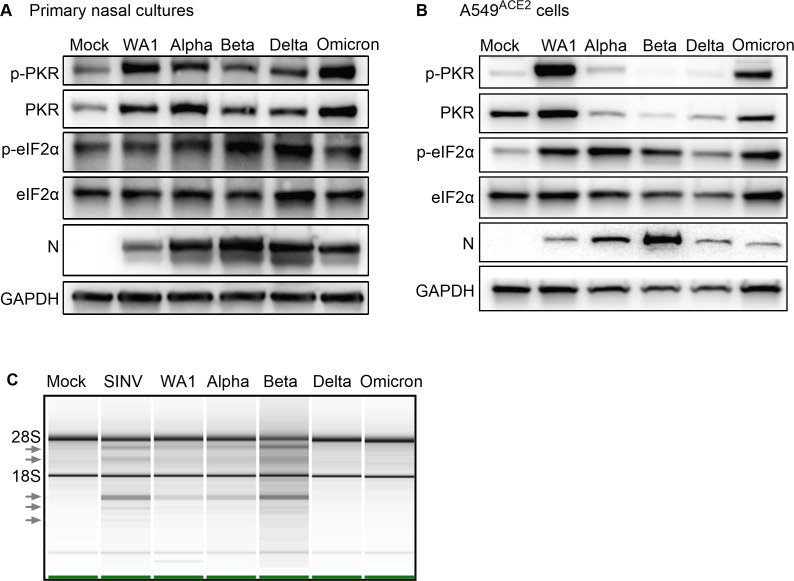
Fig 6.
dsRNA-induced pathways during SARS-CoV-2 WA1 and VOCs infections. (A) Primary nasal cultures were infected at MOI = 0.1. Cells were lysed at 96 hpi, protein extracted, and analyzed by western blot for p-PKR, PKR, p-eIF2α, and eIF2α. The controls, GAPDH and SARS-CoV-2 nucleocapsid (N), are the same as Fig. 2A. (B) A549ACE2 cells were infected at MOI = 0.1 for 72 hpi. Cells were lysed, protein extracted, and analyzed by western blot for p-PKR, PKR, p-eIF2α, eIF2α, N, and GAPDH. (A and B) Images are representative of western blots from three independent infections. (C) Infections were performed in A549ACE2 cells at MOI = 0.1. At 48 hpi (SARS-CoV-2 strains) and 24 hpi (SINV), cells were lysed, RNA extracted, and analyzed on a Bioanalyzer. Arrows indicate bands of degraded RNAs. Image is representative of two independent experiments.