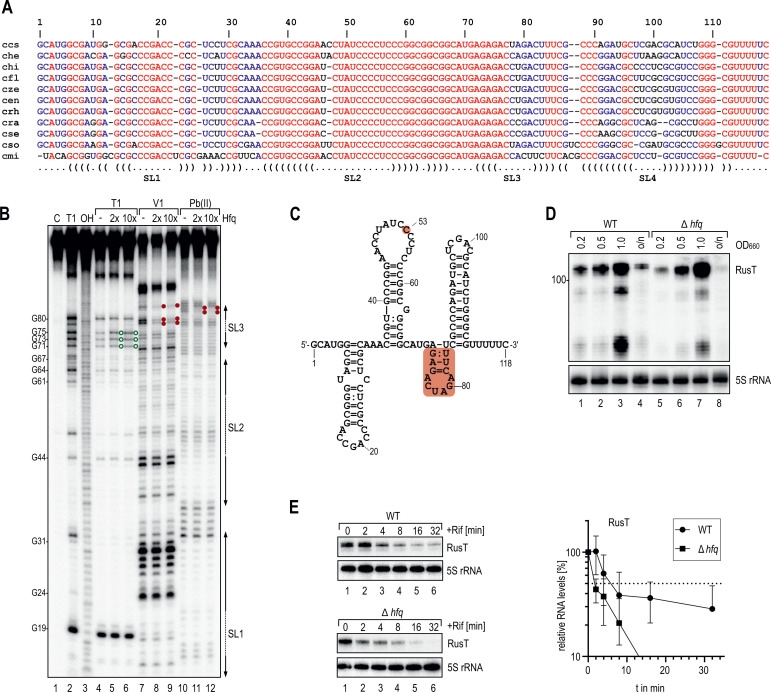
Fig 2.
Conservation, structure, and Hfq-dependence of RusT. (A) Non-redundant alignment of the RusT sRNA with orthologous sRNAs of the genus Caulobacter (ccs: C. crescentus NA1000; che: C. henricii CB4; chi: C. hibisci strain KACC 18849; cfl: C. flavus strain RHGG3; cze: C. zeae strain 410 SGCZ15; cen: C. endophyticus strain 774; crh: C. rhizosphaerae strain KCTC 52515; cra: C. radicis strain 695; cse: C. segnis strain TK0059; cso: C. soli strain Ji-3-8; cmi: C. mirabilis strain FWC 38). Colors indicate full (red); partial (blue) or no (black) conservation. The predicted secondary structure of C. crescentus RusT is indicated below the alignment. (B) In vitro structure probing of 5′ end-labeled RusT sRNA (0.4 pmol) with RNase T1 (lanes 4–6), RNase V1 (lanes 7–9), and lead(II) (lanes 10–12) in the presence and absence of purified Hfq protein (0, 0.8 pmol or 4 pmol, respectively). RNase T1 and alkaline (OH) ladders of RusT were used to map cleavage fragments, and positions of mapped G-residues are marked relative to the TSS. Increased cleavage is marked with green open circles, reduced cleavage is marked with red filled circles. (C) Secondary structure of RusT as determined by chemical probing shown in (B). The sRNA region interacting with Hfq (nt 73–84) and residue C53 are highlighted in red. (D) Northern blot analysis of RusT expression in C. crescentus wild-type and an isogenic hfq mutant strain. RNA was collected at the indicated optical density (OD660) or after 24 h of growth (o/n) in PYE medium. 5S rRNA served as loading control. (E) Determination of RusT stability. Northern blot analysis of RNA obtained from wild-type or Δhfq cells grown in minimal M2G medium to exponential phase (OD660 of 0.5). Total RNA samples were collected at indicated time points prior to, and after addition of rifampicin to inhibit transcription. The time point at which 50% of RusT had been decayed (dashed line) was calculated to determine RNA half-life. Error bars indicate the standard deviation of three independent biological replicates.