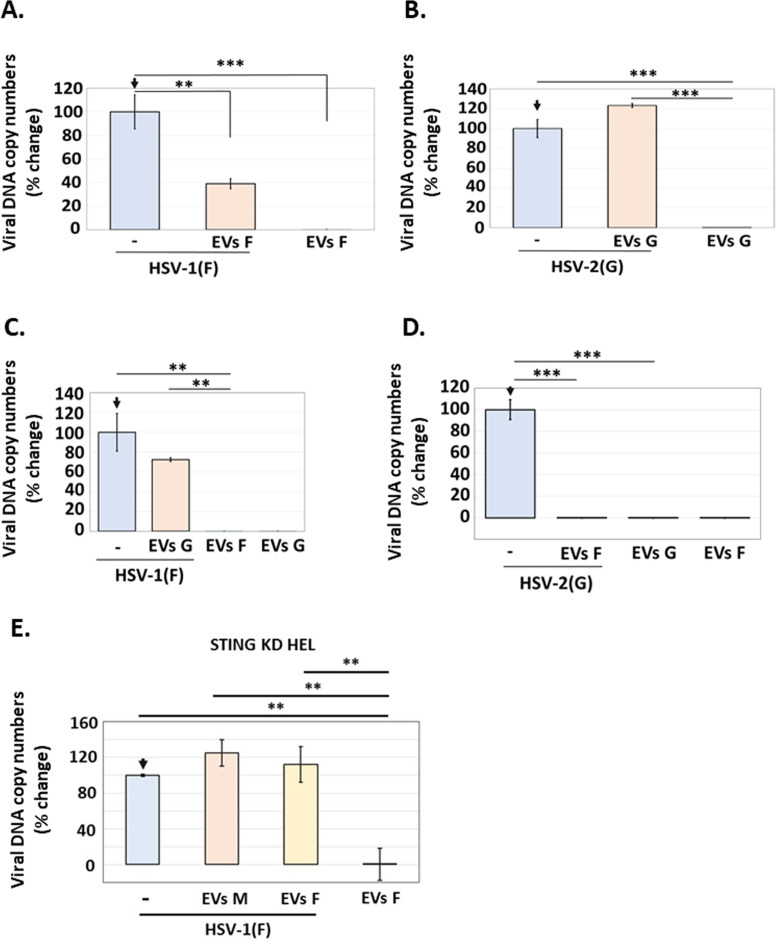
Fig 8.
EVs from HSV-1(F)-infected cells inhibited infection by HSV-1(F) or HSV-2(G), while EVs from HSV-2(G)-infected cells had no effect on HSV-1(F) or HSV-2(G). (A and B) EVs from HSV-1(F)- (A) or HSV-2(G)- (B) infected HEL cells (0.1 PFU/cell) were harvested at 24 h post-infection. EVs were separated from virus using the discontinuous sucrose-iodixanol gradient previously developed by our laboratory (29–31). Then, fractions 4–8, previously identified by our lab to contain CD63 + EVs, were collected, and EVs numbers were quantified by NTA. An equal volume of EVs was then used to expose HEL cells, corresponding to approximately 2,000 EVs/cell from HSV-1(F) and 3,000 EVs/cell from HSV-2(G). At 2 h post-EV exposure, cells were either left uninfected or infected with HSV-1(F) (A) or HSV-2(G) (B) at 0.01 PFU/cell. The cells were harvested at 24 h post-infection, and quantification of the viral genome was done by qPCR analysis as detailed in the Materials and Methods. (C and D) EVs were harvested and quantified from HSV-1(F)- or HSV-2(G)-infected HEL cells as described in A and B. HEL cells were then pre-exposed to EVs from HSV-1(F) or HSV-2(G) for 2 h, and then, cells were either left uninfected or infected with HSV-1(F) (0.01 PFU/cell) (C) or HSV-2(G) (0.01 PFU/cell) (D) to assess the effect of HSV-2(G) EVs on HSV-1(F) infection (C) or the effects of HSV-1(F) EVs on HSV-2(G) infection (D). At 24 h post-infection, the cells were harvested, and quantification of the viral genome was done by qPCR analysis as detailed in the Materials and Methods. (E) EVs were harvested and quantified from HSV-1(F)-infected STING KD HEL cells as described in A and B. Equal volume of EVs was then used to expose HEL cells, corresponding to approximately 2,000 EVs/cell from HSV-1(F) and 1,000 EVs/cell from uninfected cells. At 2 h post-EV exposure, cells were either left uninfected or infected with HSV-1(F) at 0.01 PFU/cell. The cells were harvested at 24 h post-infection, and quantification of the viral genome was done by qPCR analysis as detailed in the Materials and Methods. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.