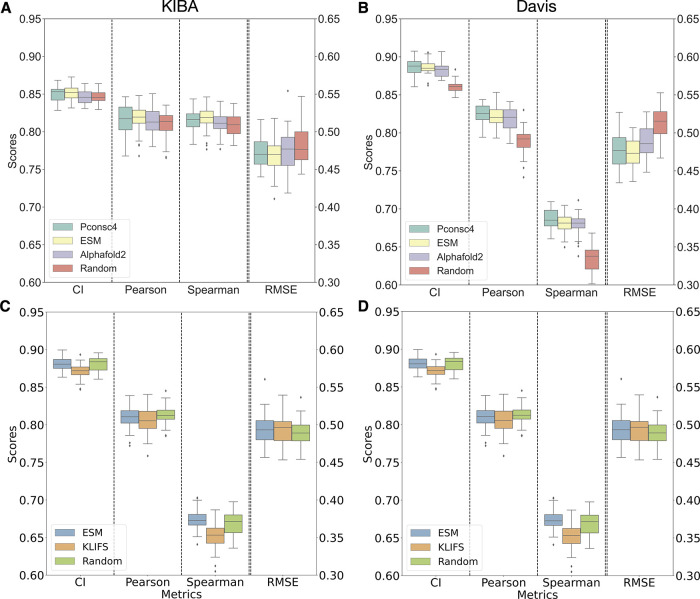
Figure 4.
Protein encodings (2D) with structural information from contact maps do not have much effect on binding affinity prediction, and 1D encodings from protein language models (PLM) perform similarly to contact maps enabled encodings. A, B: Boxplots for different performance measures (CI, Pearson correlation, Spearman Rank, and root-mean-square error) of binding affinity predictions for the KIBA data set (A) and Davis data set (B) for four different protein contact map methods. This shows that the structural information from protein contact maps encoded into a graph is not making any significant contribution to DL model performance. C, D: Boxplots for three different 1D encoding methods and their performance metrics (CI, Pearson correlation, Spearman Rank, and root-mean-square error), the PLM encodings of the ESM-1b model perform better than one-hot encodings from KLIFS handcrafted sequences on both the data sets and are comparable to random encodings. Overall, the performance of 1D encodings is comparable to the encodings that include information from protein graphs.