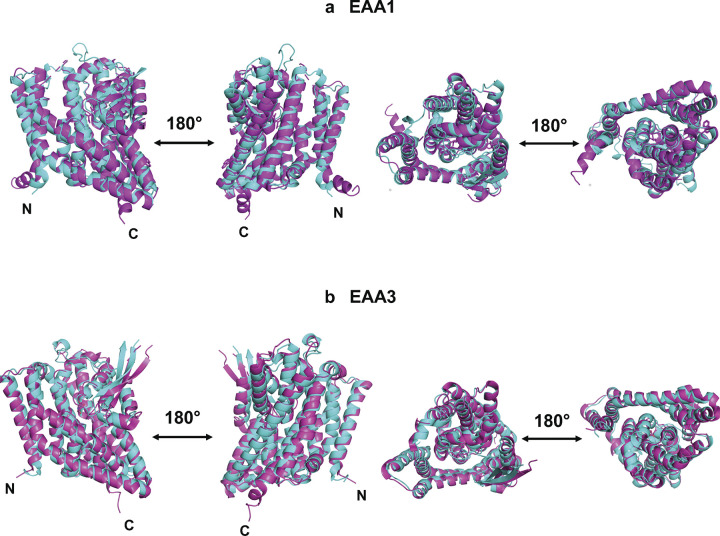
Fig 2. Superposed cryo-EM and crystal structures of EAA1Crystal and EAA3CryoEM with AlphaFold2 predicted QTY water-soluble variant and EAA1QTY and EAA3QTY.
The structures of native EAA1 (PDB ID: 5LLM, 3.25Å) and EAA3(PDB ID: 6X2Z, 3.03Å) are obtained from the Protein Data Bank. N- and C-termini are labelled. (a) The EAA1Crystal (magenta) is superposed with AlphaFold2 predicted water-soluble variant EAA1QTY (cyan). The RMSD is 1.729Å. (b) The Cryo-EM structure EAA3CryoEM (magenta) is superposed with AlphaFold2 predicted water-soluble variant EAA3QTY (cyan). The RMSD is 2.456Å. N- and C termini and large loops that are not resolved in the experimental structures are removed for clarity of direct comparisons.