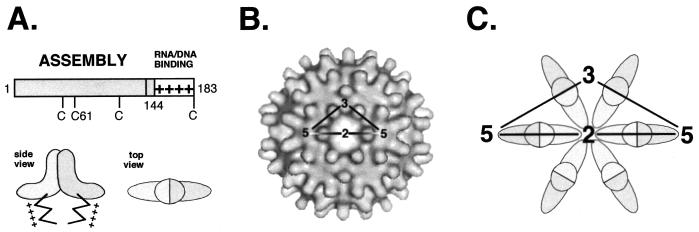
FIG. 1.
Structural organization of the HBV core protein. (A) Functional domains. The bar represents the primary sequence. The assembly domain is located in the first 144 aa and is followed by a nucleic acid binding region containing four clusters of R residues (indicated by +). The schematic representations of the core protein dimer shown below are based on cryo-electron microscopic reconstructions of capsids. The dimer interface forms the spikes visible on the capsid surface, and the R-rich C termini face its interior. (B) Architecture of the T=4 HBV capsid. A total of 120 dimers are arranged on an icosahedrally symmetric surface lattice; two-, three- and fivefold symmetry axes are indicated (adapted from reference 18). (C) Schematic representation of the arrangement of dimers around a local sixfold (strict twofold) axis of symmetry. The view is the same as in panel B.