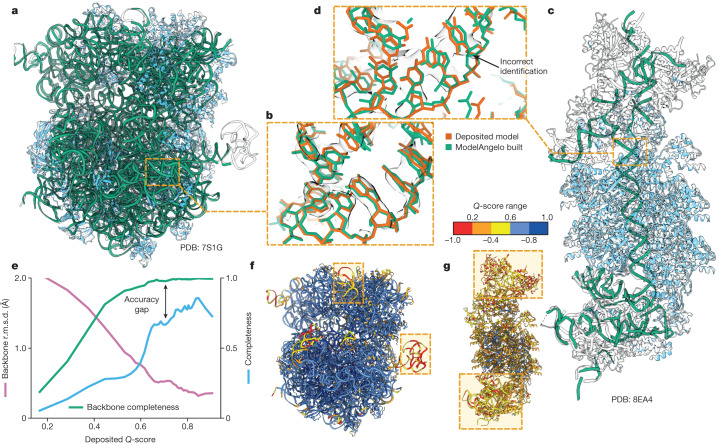
Fig. 3. Performance of ModelAngelo for nucleic acids.
a, Escherichia coli ribosome built by ModelAngelo (with ribosomal RNA in green and proteins in blue) compared with the deposited model (PDB: 7S1G, black outline)52. b, Magnified view with nucleotide bases showing high accuracy compared with the deposited model (orange). c, ModelAngelo model of the V-K CAST transpososome from S. hofmanni compared with the deposited model (PDB: 8EA4)42. Sections that were not built by ModelAngelo (black outline) are in regions of low Q-score (as shown in g). d, Magnified view comparing the nucleotide bases of both models, showing a sequence that was incorrectly identified by ModelAngelo. e, Backbone r.m.s.d., backbone completeness and sequence completeness were plotted against the deposited Q-score for six ribosome structures. f,g, Deposited models for the structures in a and c, respectively, coloured by Q-score, with low-Q-score regions indicated by boxes.