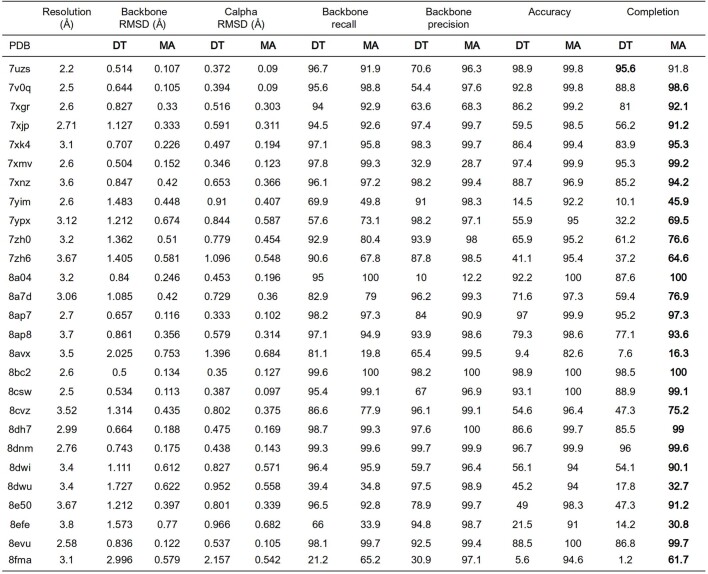
Extended Data Table 1.
Comparison with alternative approaches for the automated building of proteins
MA stands for ModelAngelo and DT for DeepTracer. Calpha RMSD is the root mean squared deviation of the predicted CA atoms against that of the deposition. Backbone RMSD is similar, but for the CA, C, O and N atoms of the protein backbones. Backbone recall is the fraction of the deposited residues that were predicted to be within 3 Å (as measured between CA atoms). Backbone precision is the fraction of the predicted residues that have a corresponding residue present in the deposition within 3 Å. Amino acid accuracy is the fraction of the predicted residues that have a correctly predicted amino acid identity. Finally, completeness is the fraction of deposited residues that were predicted with the correct base annotation. Numbers indicated in boldface are the best in each metric.