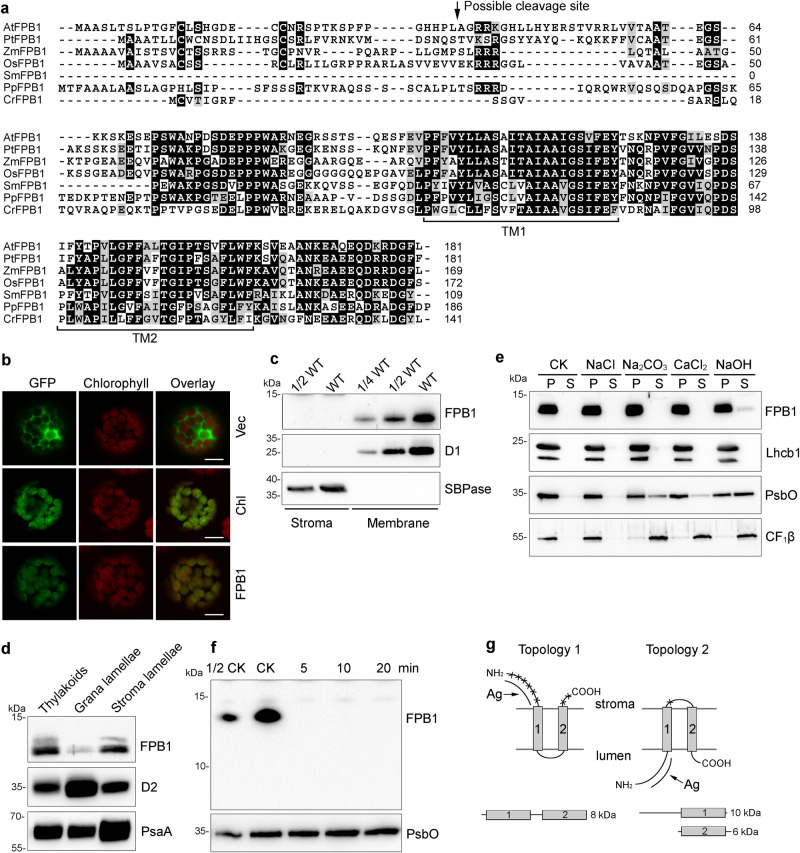
Fig. 4. Characterization of the FPB1 Protein.
a Sequence alignment of FPB1 and its orthologs from several photosynthetic eukaryotes. The two predicted transmembrane domains (TM1 and TM2) are indicated. The putative cleavage site for the chloroplast transit peptide of AtFPB1 is indicated by an arrow. b Localization of FPB1 in Arabidopsis protoplasts. Vec, empty vector control expressing only GFP protein. Chl, chloroplast localization control expressing RbcS-GFP fusion protein. FPB1, FPB1-GFP fusion protein. Bars = 10 µm. c Immunoblot analysis of FPB1. Intact chloroplasts isolated from WT plants were fractionated into stromal and membrane fractions, which were further analysed by immunoblotting with FPB1 antiserum. Antibodies against D1 and SBPase (fructose−1,6-bisphosphatase) were used as controls. d Localization of FPB1 in thylakoids. Thylakoids were fractionated into grana and stroma lamellae and immunoblot analysis was performed using antibodies against FPB1, D2, and PsaA, respectively. e Salt and alkali washing of thylakoid membranes. WT thylakoids were incubated without (CK) or with 0.25 M NaCl, 0.2 M Na2CO3, 1 M CaCl2, and 0.1 M NaOH for 30 min on ice. Soluble and membrane fractions were separated by centrifugation and immunoblotted with FPB1, Lhcb1 (integral membrane marker), PsbO (peripheral membrane protein marker exposed to the lumenal side of thylakoids), and CF1β (peripheral membrane protein marker exposed to the stromal side of thylakoids) antisera. f Trypsin digestion analysis. Thylakoids were treated with 10 μg/mL trypsin for 0 (CK), 5, 10, and 20 min on ice. The proteins were then probed with antibodies against FPB1 and PsbO (lumenal proteins that are not accessible to the enzyme), respectively. g Two possible topologies of FPB1 in thylakoids. Cleavage sites in FPB1 by trypsin are indicated by asterisks. Ag represents the FPB1 region used to raise the antibody. A total of three proteolytic fragments in the two topologies are indicated but only the 10-kDa fragment with Topology 2 can be detected with FPB1 antiserum. SDS-urea-PAGE (c–e, PsbO in f) and Tricine-SDS-PAGE (FPB1 in f) were used to separate proteins. Data are representative of two independent biological replicates (b–f). Source data are provided as a Source Data file.