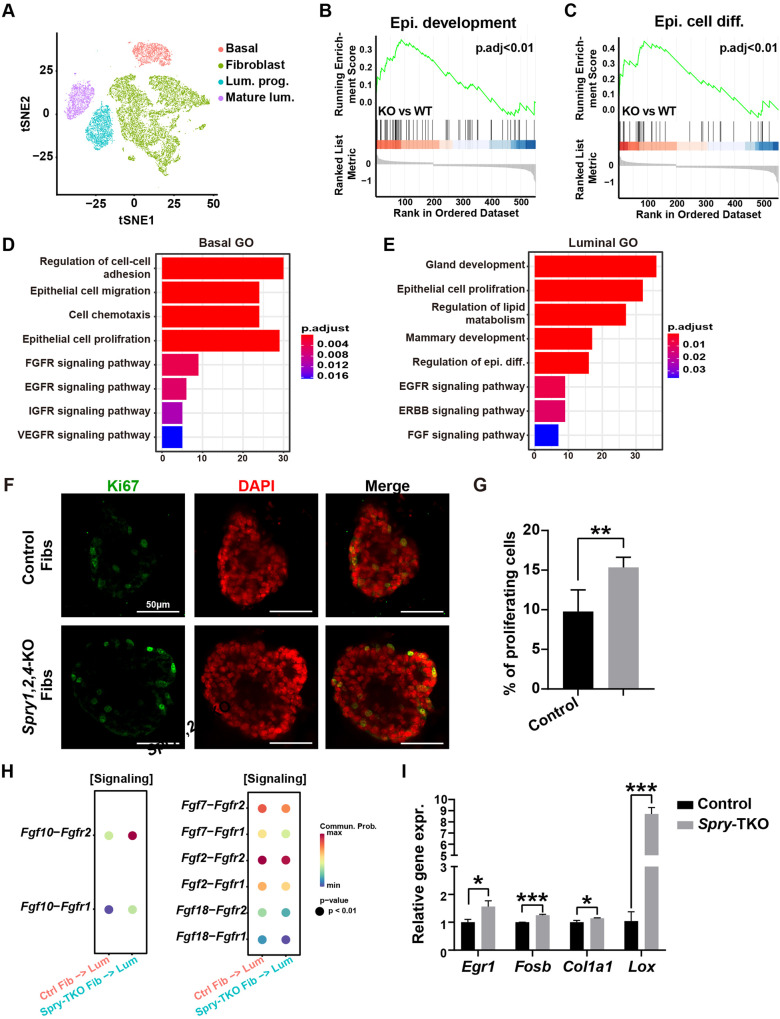
Fig. 2. Loss of Spry genes in mammary fibroblasts leads to increased FGF10 but decreased FGF2 signaling in the epithelium.
A t-SNE plot showing the combined single cell transcriptomes of mammary epithelial cells and stromal fibroblasts isolated from seven-week-old control and Spry-TKO mammary glands (n = 2 female mice). Abbreviations: Lum, luminal; prog, progenitor. B, C GSEA analysis of pathways related to epithelial development and cell differentiation in control and Spry-TKO mammary glands. Abbreviations: diff., differentiation; epi., epithelial; KO, knockout; WT, wildtype. D, E GO analysis of the main pathway changes in basal cells (D) and luminal cells (E) of control and Spry-TKO mammary glands. F, G Cell proliferation of wild-type organoid epithelium co-cultured with control and Spry1,2,4-KO fibroblasts (Fibs) as detected by Ki67 staining (F) and was quantified (G). Data were from three independent experiments and were presented as mean ± SD. **P < 0.01. Scale bars: 50 μm. H Cellchat analysis of either increased or decreased fibroblast-to-epithelial FGF ligand-receptor signaling in Spry-TKO mammary glands when compared with control glands. Abbreviations: ctrl., control; fib., fibroblasts; lum., luminal. I qPCR analysis of target genes in the FGF signaling pathway. Abbreviation: Expr, expression. *P < 0.05; ***P < 0.001.