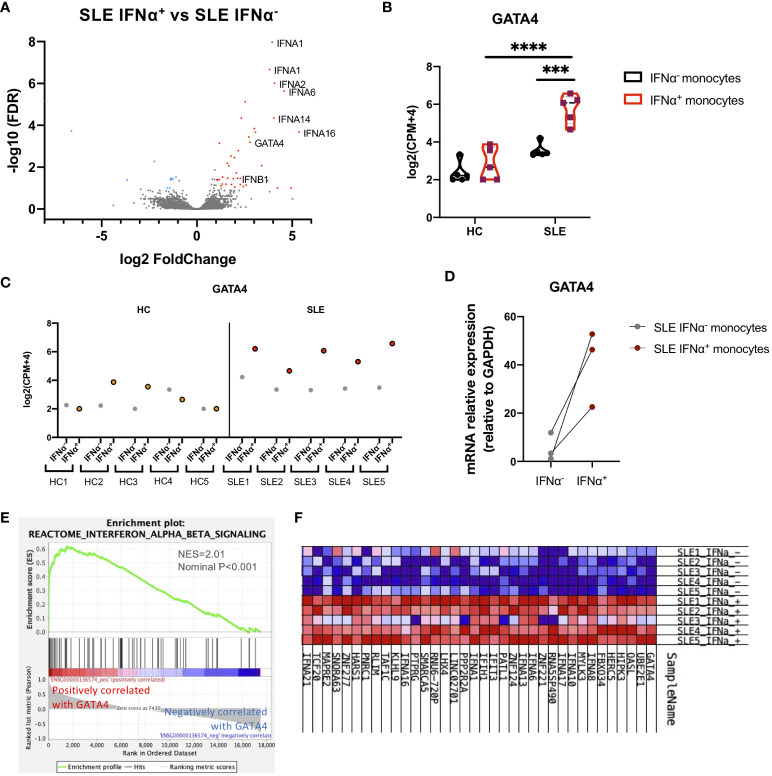
Figure 2.
GATA4 is upregulated in IFNα-producing SLE monocytes. RNA-seq analysis of IFNα positive/negative SLE and HC monocytes in the experiments of Figure 1 . (A) A volcano plot comparing the FDR and fold-change of total gene expression in RNA-seq data of IFNα+ and IFNα− SLE monocytes. (B) A violin plot showing GATA4 gene expression levels in RNA-seq data of IFNα+ and IFNα− monocytes from SLE or HC. Each dot indicates the log2 transformed gene expression level of GATA4 (mean ± SEM, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test. ***P<0.001 and ****P<0.0001). (C) GATA4 gene expression levels in RNA-seq data from individual samples are plotted. IFNα− = IFNα− monocytes, IFNα+ = IFNα+ monocytes. (D) Gene expression of mRNA GATA4 was quantified by real-time reverse transcription-quantitative PCR (RT-qPCR) in IFNα− and IFNα+ SLE monocytes. (E, F) GSEA analysis using GATA4 (ENSG00000136574) as a phenotype gene was performed in RNA-seq data of IFNα+ and IFNα− SLE monocytes. (E) GSEA plot showing the enrichment of “Interferon Alpha Beta Signaling (R-HSA-909733)” in genes that have a positive correlation with GATA4 expression. (F) Expression profiles of genes with expressions that were positively correlated with GATA4 expression. Heatmap red-blue color intensity indicates the expression level of genes (red, high; white, average; blue, negative).