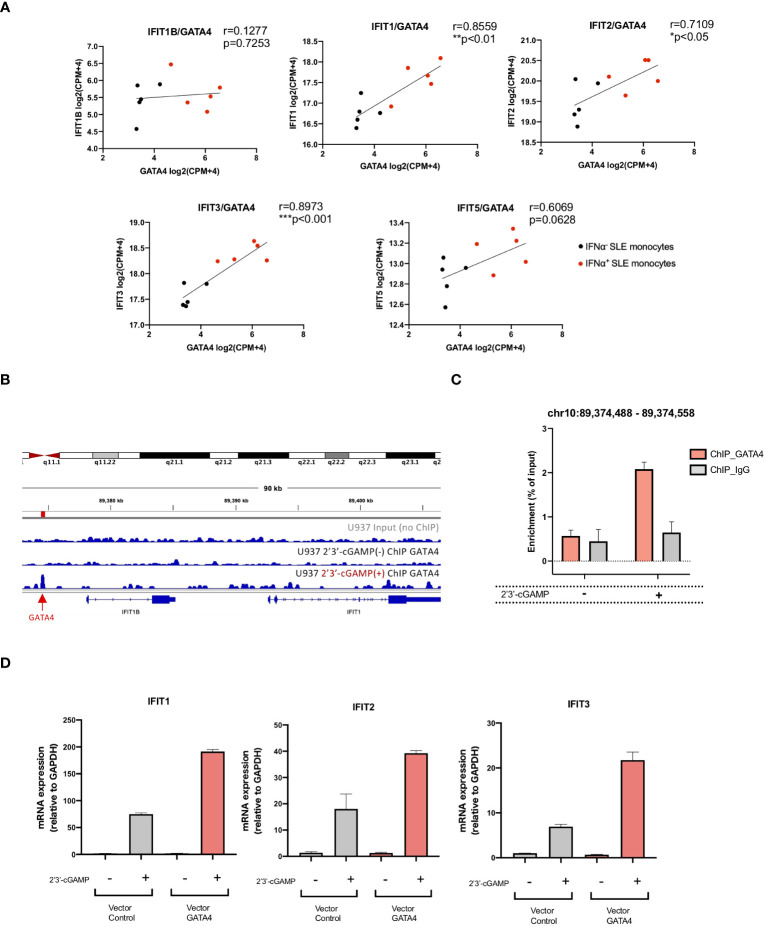
Figure 5.
GATA4 regulates IFIT family gene expression through the GATA4-bound enhancer element. (A) The correlation between the gene expressions of GATA4 and IFIT family genes in RNA-seq data for IFNα+ monocytes and IFNα− monocytes from SLE was assessed by Pearson’s correlation analysis (*P<0.05, **P<0.01, and ***P<0.001). (B) GATA4 chromatin immunoprecipitation (ChIP)-seq analysis in U937 cells with or without 2′3′-cGAMP stimulation was performed. Genome Browser image of the region upstream of IFIT1B and IFIT1. The arrow and red bar indicate the predicted GATA4-binding site. (C) GATA4 ChIP was performed in U937 cells with or without 2′3′-cGAMP stimulation. GATA4 binding at the genomic region (chr10: 89,374,488 – 89,374,558) was determined by ChIP-qPCR. Results are representative of three independent experiments (mean ± SEM of triplicate RT-qPCR measurements). (D) U937 cells transfected with a GATA4 expression vector or empty vector control were stimulated with 2′3′-cGAMP. Expressions of IFIT1, IFIT2, and IFIT3 were quantified by RT-qPCR. The 2-ΔΔCT method was used to calculate the relative gene expression, with normalization to GAPDH as the internal control. Results are representative of three independent experiments (mean ± SEM of triplicate RT-qPCR measurements).