Abstract
An arginine-rich peptide from the Jembrana disease virus (JDV) Tat protein is a structural “chameleon” that binds bovine immunodeficiency virus (BIV) or HIV TAR RNAs in two different binding modes, with an affinity for BIV TAR even higher than the cognate BIV peptide. We determined the NMR structure of the JDV Tat–BIV TAR high-affinity complex and found that the C-terminal tyrosine in JDV Tat forms a network of inter- and intramolecular hydrogen bonding and stacking interactions that simultaneously stabilize the β-hairpin conformation of the peptide and a base triple in the RNA. A neighboring histidine also appears to help stabilize the peptide conformation. Induced fit binding is recurrent in protein–protein and protein–nucleic acid interactions, and the JDV Tat complex demonstrates how high affinity can be achieved not only by optimization of the binding interface but also by inducing new intramolecular contacts that stabilize each binding partner. Comparison to the cognate BIV Tat peptide–TAR complex shows how such a costabilization mechanism can evolve with only small changes to the peptide sequence. In addition, the bound structure of BIV TAR in the chameleon peptide complex is strikingly similar to the bound conformation of HIV TAR, suggesting new strategies for the development of HIV TAR binding molecules.
Keywords: NMR, RNA structure, RNA-binding domain
The ability of macromolecules to interact with high affinity and specificity is often accompanied by conformational changes in the binding partners (1–4). Flexibility of one or both molecules can contribute to optimization of the binding surfaces and allow binding to multiple partners (5, 6). Numerous examples of conformational adaptability have been observed in protein–protein and protein–nucleic acid interactions, including Ig proteins, which can accommodate a remarkably wide range of binding partners, and the ribosome, where induced-fit binding helps direct its ordered assembly (7–9).
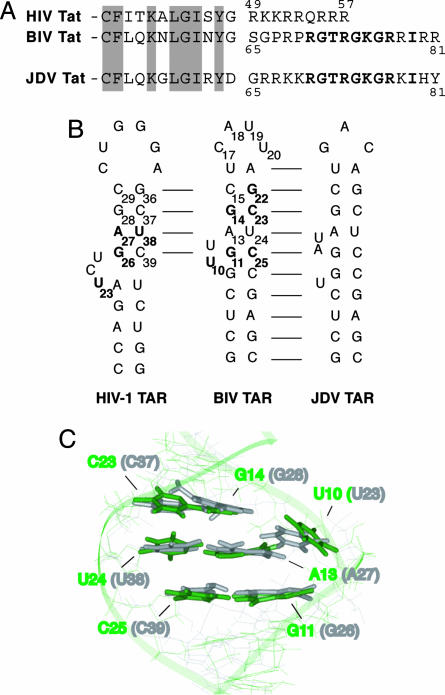
In RNA–protein interactions, the arginine-rich motif (ARM) has served as a model system for examining structural mechanisms that underlie induced fit binding (10–12). Studies of ARM peptide–RNA complexes, including the Tat ARM–TAR RNA interactions of several lentiviruses, have shown that the unbound ARMs generally are unfolded and can adopt a variety of conformations upon RNA binding, often with a concomitant change in RNA structure. In the case of HIV Tat, a relatively weak binding ARM peptide remains in an extended conformational when bound to HIV TAR but causes a large conformational change in the RNA, inducing stacking between the two helical stems and formation of a U–A:U base triple (13–23). In the case of bovine immunodeficiency virus (BIV) Tat, a conformational change in BIV TAR also is observed upon binding, but the cognate ARM peptide undergoes a large conformational rearrangement, forming a β-hairpin structure that facilitates high-affinity binding through a large set of specific contacts to the RNA (24–29). Despite the striking difference in binding modes of these two ARM peptides, HIV and BIV TAR RNAs have closely related sequences and secondary structures (Fig. 1B), and their bound three-dimensional conformations around the bulge regions are very similar (Fig. 1C) (30). Even with these similarities, the BIV Tat ARM cannot adopt the high-affinity β-hairpin conformation with HIV TAR.
Fig. 1.
Comparison of Tat ARM domains and TAR RNAs. (A) The HIV-1, BIV, and JDV Tat ARM domains are aligned based on homology between the N-terminal activation domains (partially shown, with conserved residues shaded). Analogous residues in the BIV and the JDV Tat ARM are shown in bold. (B) Secondary structures of the HIV-1, BIV, and JDV TAR hairpins. Sequence identity is indicated by the lines, and nucleotides in HIV-1 and BIV TARs important for binding by the cognate protein are shown in bold. (C) Superimposition of the Tat binding sites in BIV TAR (green) and HIV-1 TAR (gray) in their bound conformations, from NMR models of the HIV-1 TAR-arginamide (19) and BIV Tat–TAR (28) complexes. The structures were superimposed by using all bases known to be important for binding of each, and the corresponding base numbering is shown for both RNAs.
We previously described a “chameleon” Tat peptide from Jembrana disease virus (JDV) (31, 32) that is related to the BIV ARM and is able to bind not only to its cognate JDV TAR but also to HIV and BIV TARs (33). This ARM domain is particularly interestingly for two reasons: first, it behaves as a structural chameleon that binds to HIV and BIV TARs in the HIV Tat and BIV Tat respective binding modes and, second, it binds BIV TAR with even higher affinity than the cognate BIV peptide because of amino acid differences at its C terminus (33). Indeed, replacing C-terminal residues of the BIV ARM with those of JDV increases affinity for BIV TAR by ≈10- to 60-fold depending on the nature of the N-terminal residues, largely because of a C-terminal Tyr residue in the JDV ARM (33). Thus, the JDV chameleon provides a particularly good opportunity to investigate how one peptide can recognize several RNA targets and how one RNA target can be bound by different peptides. Here we address the latter question, presenting the NMR structure of the high affinity JDV Tat peptide–BIV TAR complex and its comparison to the cognate BIV complex. We find that amino acids in the C terminus of the JDV peptide enhance RNA-binding affinity by creating additional RNA–protein interactions and, more importantly, by stabilizing the structures of both the protein and RNA. This mechanism of costabilization highlights how intramolecular interactions can enhance the binding affinity of two flexible partners. Furthermore, we find that small changes in peptide sequence not only can alter direct contacts to the RNA but also change the induced fit binding mechanism and influence the structural and folding pathways in which specificities are evolved through costabilizing the binding partners. The stabilizing effect on BIV TAR is especially interesting because it generates a conformation nearly indistinguishable from that of HIV TAR, suggesting how side chain moieties from the JDV peptide might be used to develop high-affinity HIV TAR binding molecules.
Materials and Methods
Peptides and RNAs. JDV Tat peptides were synthesized on an Applied Biosystems Model 432A peptide synthesizer by using Fmoc chemistry and standard Applied Biosystems resin (25 μmol) and protecting groups. Peptides were capped with an acetyl group at the N terminus and an amide group at the C terminus. After cleavage and deprotection, peptide was purified on a C4 reverse-phase high-performance liquid chromatography column (Vydac 214TP54) at a flow rate of 5 ml/min by using an acetonitrile gradient (2% per min) in 0.1% trifluoroacetic acid. Molecular masses were confirmed by laser desorption mass spectroscopy on a Voyager-DE MALDI/TOF spectrometer (PerSeptive Biosystems, Framingham, MA).
TAR RNAs were prepared by T7 RNA polymerase-based in vitro transcription using synthetic DNA templates (34). All RNAs contained two additional guanine nucleotides at the 5′ end for improved transcription efficiency and two cytosines at the 3′ end to base pair with the guanines and maintain the hairpin. For gel-shift assays, in vitro transcribed RNAs were uniformly labeled by incorporating [α-32P]rCTP during transcription reactions (44 μl) incubated for 4 h at 37°C. RNAs were ethanol precipitated, purified on denaturing 15% polyacrylamide/urea gels, and resuspended in sterile deionized water. RNA concentrations were determined from the specific activity of [α-32P]rCTP incorporated into the transcripts. TAR RNAs (20 nM) were annealed by heating at 85°C for 5 min and slow cooling to room temperature in renaturation buffer (200 mM Tris·HCl, pH 7.5/1 M NaCl).
For NMR experiments, RNAs were synthesized by using DNA templates containing two 2′ methoxy guanosines at the 5′ terminus to reduce the addition of nontemplated nucleotides by the polymerase and thus ease purification and increase the yield of correct length transcripts (35). RNAs were separated on denaturing 20% polyacrylamide/urea gels, electroeluted, and ethanol precipitated. Resuspended samples were dialyzed three times over 48 h, first against 10 mM sodium phosphate, pH 6.5/100 mM NaCl/0.1 mM EDTA, then against 10 mM sodium phosphate pH 6.5/50 mM NaCl, and then against deionized water.
RNA-Binding Gel Shift Assays. Peptide and RNA (0.2 nM) were incubated together on ice for 10 min in 40 mM Hepes-KOH, pH 7.5/400 mM KCl/4 mM MgCl2/2 mM EDTA/4 mM DTT/40% glycerol, with 200 μg/ml tRNA as competitor. Samples were loaded onto prerun 10% native polyacrylamide gels and electrophoresed at 200 V for 3 h at 4°C. Gels were dried and exposed to a PhosphorImaging plate for 12 h, and bands were quantified with a Molecular Dynamics PhosphorImager and imagequant software. Binding constants were estimated by measuring the disappearance of the unbound RNA, fitting the data to binding curves by using kalei-dagraph software (Synergy Software, Reading, PA).
NMR Spectroscopy. NMR samples were prepared in 10 mM sodium phosphate, pH 6.5/50 mM NaCl in either 90%H2O/10% D2O or 100% D2O at a concentration of 1.5 mM RNA. Peptide–RNA complexes were formed by incrementally adding JDV Tat peptide to 1:1 stoichiometry, monitoring the imino proton spectra of TAR until no further changes were observed. Homonuclear 2D COSY, TOCSY, and NOESY spectra were recorded at 10°C, 15°C, and 25°C either in H2O or D2O. For spectra acquired in D2O, the residual HDO signal was suppressed by DANTE presaturation; in H2O, solvent was suppressed by using a symmetrically shifted shaped pulse (36). Two-dimensional NOESY spectra were acquired at 100-, 200-, and 400-ms mixing times. All NMR experiments were carried out on a Varian Unity Plus 600-Mhz spectrometer, and spectra were processed with nmrpipe (37) and analyzed with sparky (38).
The assignment of RNA protons was aided by comparison to the spectra of the BIV Tat–BIV TAR complex, as protons of nucleotides in the lower stem and some in the bulge region showed the same chemical shifts (28, 29). Assignments of base pair imino and most amino protons were obtained from NOESY spectra collected in H2O. Nonexchangeable protons were assigned by using a combination of DQF-COSY and NOESY in D2O, and all resonances of the nonexchangeable aromatic and H1′ protons were identified. Several H3′,H4′, and H5′/H5″ protons were assigned from NOEs to the H1′ atoms of the same sugar, where resonances were resolvable. Next, we traced the base–H1′–base proton connectivities to assign each base sequentially. The sequential assignments were confirmed by using connectivities between exchangeable imino and amino protons. Chemical shift data for the RNA are provided in Table 4, which is published as supporting information on the PNAS web site. Protons of the JDV Tat peptide were assigned as described (39). Sequential assignment of the five arginines was facilitated by previous assignments of Arg-70, Arg-73, and Arg-77 in the BIV Tat–BIV TAR complex, which showed the same chemical shifts (28, 29). Chemical shift data for the peptide are provided in Table 5, which is published as supporting information on the PNAS web site.
Distance restraints were derived from 200-ms D2O and 100-ms H2O spectra. NOE crosspeak intensities were converted into upper limit distances by using the caliba function in cyana (40), with the H5/H6 cross peaks of pyrimidines as standards. The lower limit for distance restraints was 1.8 Å. A pseudoatom correction for unassigned stereopartners and magnetically equivalent protons was applied as described (41). NOE crosspeaks present only in the 400-ms mixing time spectrum, as well as NOE crosspeaks from H2O spectra, were assigned as 3.5- to 7.5-Å distances. Hydrogen bonds between base pair atoms, as suggested by NOEs in H2O, were included as distance restraints and represented as two ranges of distances (1.8–2.0 Å for proton and hydrogen-bond acceptor and 2.7–3.0 Å for hydrogen-bond acceptor and donor atoms). All restraints that do not contain structural information, such as fixed or redundant distances and distances that cannot be violated, were removed before the calculations. For ribose endocyclic torsion angles, the presence of a strong H1′–H2′ COSY (JHNα > 8 Hz) crosspeak was interpreted as a C2′ endo conformation, whereas the absence of a crosspeak (JHNα < 3Hz) was interpreted as a C3′-endo conformation, using a 20° range around the standard values (42) as restraints. All other sugars angles were left unrestrained, and no other assumptions about RNA conformation were placed on the calculations.
Three-dimensional structures of the complex were calculated by using the program cyana on a Silicon Graphics O2 workstation. Structures were calculated beginning with 1,000 random structures and incrementally adding distance constraints in four iterations to avoid local minima. The final 30 structures with the lowest number of violations were subjected to restrained molecular dynamics (rMD) refinement using cns software (version 1.1) and standard cns topology and parameter files for RNA and protein (43). The high temperature dynamics were performed at 1,000 K with 2,500 steps over 0.006 ps in cartesian space, followed by a slow-cooling annealing stage, in cartesian space, from 300 K to 0 K in 2,000 steps of 0.004 ps. After this initial minimization, hydrogen bond restraints were added for each base pair in the stems as well as between the H3 of U10 and N7 of A13, as deduced by the presence of a U10 imino proton resonance (see Results and Discussion). Eleven planarity restraints were introduced for each base pair, and 61 dihedral restraints were used to constrain the sugar puckers as C2′-endo or C3′-endo for distance geometry calculations. Force constants for NOE distance and dihedral restraints were maintained at 150 kcal·mol–1·Å–2 and 20 kcal·mol–1·rad–2, respectively. For the final minimization, the NOE force constant was maintained at 75 kcal·mol–1·Å–2 with a force constant of 150 kcal·mol–1·Å–2 to maintain base pair planarity. Structures were superimposed, rms deviations were calculated by using molmol (44), and calculated molecules were visualized by using insight ii (Molecular Simulations, Waltham, MA).
Results and Discussion
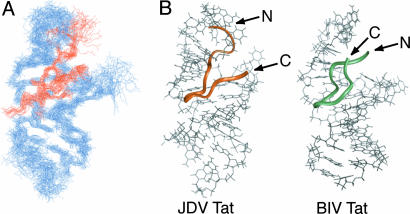
Overall Structure of JDV Tat Peptide–BIV TAR Complex. To understand how BIV TAR is recognized with high affinity by the JDV Tat peptide (residues 65–81) and to compare its binding mode to that of BIV Tat, we determined the structure of the JDV ARM peptide–BIV TAR complex by using 2D proton NMR spectroscopy. The BIV TAR hairpin (nucleotides +4to +31 of the mRNA) was identical to that used in determining the structure of the BIV Tat–TAR complex (28, 29), which aided in assigning the spectra. A total of 761 distance constraints, including 183 intermolecular constraints (intermolecular NOEs are provided in Table 6, which is published as supporting information on the PNAS web site) and 61 NMR-derived dihedral angle constraints, was used to calculate the structure using the torsion-space molecular dynamic (cyana) and restrained energy minimization (cns) (Tables 1, 2, 3). The 10 lowest-energy structures had few NOE violations and superimposed with an average pairwise rms deviation of 1.59 Å for the peptide and RNA backbone atoms and 1.79 Å for all heavy atoms. Residues 70–81 of the peptide and the stems and U10 bulge region of the RNA (nucleotides 5–11, 13–16, and 21–30) are particularly well defined over the ensemble of structures (Fig. 2A and Tables 1, 2, 3).
Table 1. Distance and dihedral angle constraints for JDV Tat—BIV TAR complex.
| Peptide | RNA | Complex | |
|---|---|---|---|
| Total number of distance constraints | 293 | 226 | 700 |
| Intraresidue | 114 | 81 | |
| Sequential | 54 | 90 | |
| Medium/long range | 125 | 55 | |
| Hydrogen bonds | 61 | ||
| Peptide—RNA | 181 | ||
| Dihedral angle constraints | 61 |
Table 2. Structure statistics of 10 final structures for JDV Tat—BIV TAR complex.
| NOE violations, number >0.2 Å | 7.7 ± 4.3 |
| Angle violations, number >2° | 3.1 ± 0.5 |
| Mean deviation from ideal covalent geometry | |
| Bond length, Å | 0.0032 ± 0.0001 |
| Bond angle, ° | 0.616 ± 0.019 |
| Impropers, ° | 0.426 ± 0.029 |
Table 3. Average pairwise rms deviations, in Å, for JDV Tat—BIV TAR complex.
| Backbone | Heavy | |
|---|---|---|
| Complex | ||
| All residues | 1.59 ± 0.33 | 1.79 ± 0.37 |
| Peptide residues 70-81; RNA nts 5-11, 13-16, and 21-30 | 1.22 ± 0.29 | 1.21 ± 0.21 |
| Peptide | ||
| All residues | 0.99 ± 0.11 | 1.44 ± 0.28 |
| Residues 70-81 | 0.78 ± 0.17 | 1.12 ± 0.17 |
| RNA | ||
| Stems with U10 bulge (nt 5-11, 13-16, and 21-30) | 1.32 ± 0.24 | 1.24 ± 0.22 |
| Stems, U10 and Loop (nt 5-11 and 13-30) | 1.51 ± 0.30 | 1.50 ± 0.29 |
nt, nucleotides.
Fig. 2.
Overall structure of the JDV Tat ARM–BIV TAR complex. (A) Superposition of the 10 lowest energy minimized structures on the minimized average structure. Only heavy atoms were used for the superposition and are shown. (B) Comparison of the minimized average structures of the JDV Tat ARM (residues 65–81)–BIV TAR complex (Left) and BIV Tat ARM (residues 68–81)–BIV TAR complex (28) (Right). The structures, as seen from the major groove, show the peptides as colored ribbons and BIV TAR in gray. The N and C termini are indicated but poorly ordered in the BIV Tat complex.
As anticipated from the sequence similarity and preliminary NMR experiments (33), the JDV Tat ARM generally binds BIV TAR in a manner similar to the BIV Tat ARM. The peptide, which is unstructured in the absence of RNA (data not shown), inserts deeply into the RNA major groove and adopts a β-ribbon-like conformation, with two antiparallel strands (residues 71–73 and 77–79) linked by a sharp turn (residues 74–76) (Fig. 2B). Unlike the BIV Tat ARM, the N- and the C-terminal regions of the JDV ARM (residues 65–69 and 80–81) are in close proximity to the RNA, although they do not adopt particular protein secondary structures.
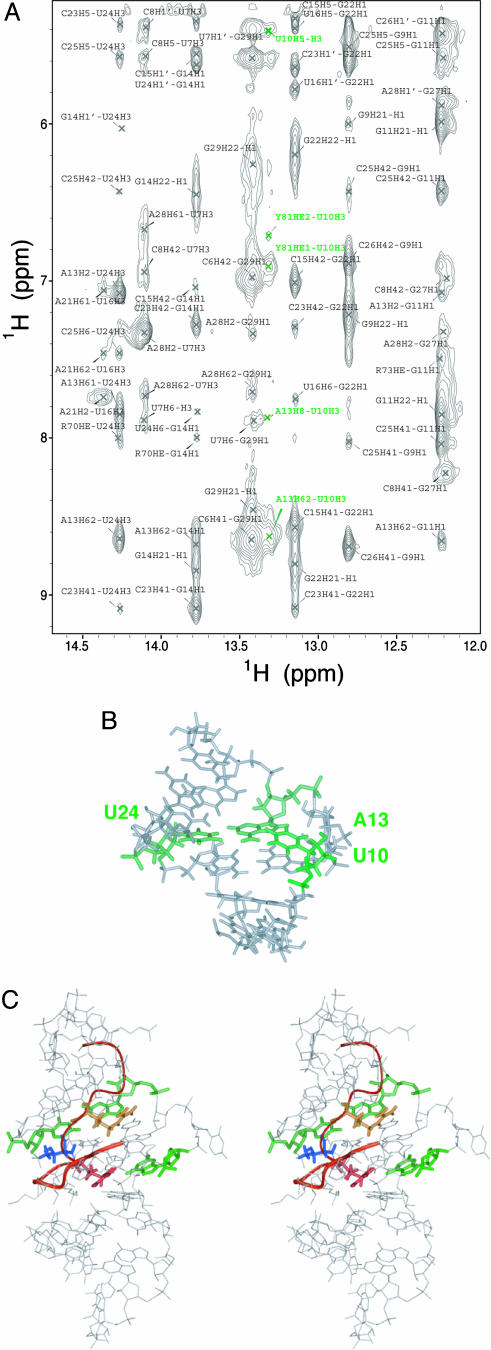
The overall structure of BIV TAR, particularly the stem and bulge regions, is quite similar between the JDV and BIV peptide complexes (Fig. 2B). The two stems coaxially stack, as indicated by sequential NOEs between protons of G11, G9, and U24, generating a nearly contiguous A-form helix. The two bulged nucleotides (U10 and U12) are not intercalated into the helix but induce a minor distortion that locally enlarges the major groove (to 15.6 Å versus 10.5 Å in an A-form helix) and allows insertion of the peptide. The U12 base is solvent exposed and disordered, whereas U10 is positioned in the major groove near the A13:U24 base pair and gives a strong imino proton resonance (Fig. 3A) indicative of a stable hydrogen bond and consistent with formation of a base triple (Fig. 3B). The presence of the U10 imino resonance under our acquisition conditions (pH 6.5, 10°C) was surprising given that it only could be seen under relatively extreme conditions (pH 5.5, –2°C) in the BIV peptide complex (28, 29). Thus, the base triple in BIV TAR appears significantly more stable in the context of the JDV peptide. Unlike in the BIV ARM complex, the loop structure is well defined, although the rms deviations of the four nucleotides composing the loop is higher than for the stems and U10 bulge region (2.43 ± 0.76 Å and 1.32 ± 0.24 Å for all heavy atoms, respectively). Although the C17, A18 and U20 bases point toward the major groove, the U19 base is flipped out, probably to accommodate the large side chains of arginines 66 and 67.
Fig. 3.
Interactions within the JDV Tat ARM–BIV TAR complex. (A) Imino region of the JDV Tat ARM–BIV TAR complex NOESY spectrum. The spectrum was recorded in H2O at 10°C, pH 6.5. Cross peaks labeled in green correspond to intra- and intermolecular NOEs between the imino proton of the U10 bulge residue and protons of U10 and Y81 and A13. (B) Major groove view of the minimized average structure highlighting the base triple formed between U10, A13, and U24 (green). For clarity, the peptide and RNA loop are not shown. (C) Stereoview of the minimized average structure showing the interactions also found in the BIV Tat–TAR complex: Arg-70 (yellow) is in proximity to G11 (green), Arg-73 (red) is in proximity to G14 (green), and Thr-72 (blue) contacts C23 (green).
JDV and BIV Tat Core Domains Contact the RNA in a Similar Way. The core regions of the JDV and BIV ARM peptides, defined as residues 70–77, have nearly identical sequences (Fig. 1 A) and adopt the same backbone conformation and make similar contacts to the RNA (Fig. 3C). Even though no exceptional electrostatic constraints were imposed in our calculations, almost all structures place the Hε and ηNH2 atoms of the Arg-70 and Arg-73 guanidinium groups within hydrogen bonding distance of the O6 and N7 atoms of G14 and G11, respectively, as observed in the BIV Tat–TAR complex (28, 29). In all models, Thr-72 is positioned to form a pair of hydrogen bonds to nucleotide C23. The observed resonance of the threonine hydroxyl proton, also observed in the BIV complex spectra, suggests a hydrogen bond to the phosphate oxygen of C23, and the backbone carbonyl is positioned to hydrogen bond to the amino group of the base. Gly-71 and Gly-74 do not interact directly with the RNA but, as in the BIV ARM complex, they sterically allow deep insertion of the peptide into the major groove. Both glycines adopt positive φ angle values that are energetically unfavorable for other amino acids, allowing formation of the sharp turn. All of the interactions observed with the core residues are consistent with the critical requirement of each in the BIV peptide interaction (27).
Roles of the Peptide Termini. We identified 183 intermolecular constraints for the JDV complex compared to 104 for the BIV complex (29), with the majority of additional constraints (71%) coming from the peptide termini. As a consequence, the N- and C-terminal segments of the JDV peptide (residues 65–69 and 80–81) are in intimate contact with the RNA, in contrast to the unconstrained termini observed in the BIV complex, which do not contact the RNA and are highly exposed to solvent (Fig. 2B). The JDV N-terminal segment, which is highly basic (Fig. 1A), is packed against the loop but makes no specific contacts. Despite the more ordered N terminus and RNA loop compared to the BIV complex, previous mutagenesis experiments showed that replacing the N-terminal residues of the JDV ARM with those of BIV actually enhanced binding affinity (33). Thus, it appears that any enthalpy gained from these largely electrostatic contacts is more than offset by the entropic costs of conformational changes to the peptide N terminus and RNA loop.
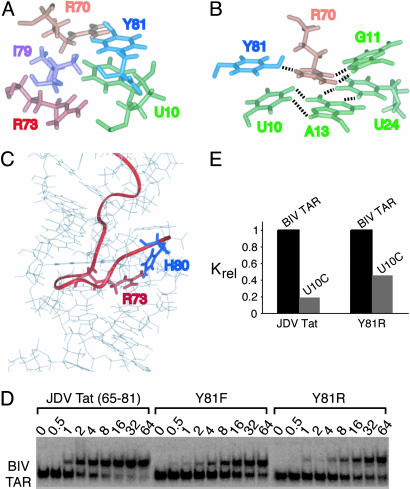
In contrast, the C-terminal tyrosine and histidine residues are responsible for the increased affinity for BIV TAR (33), which can be explained by the extensive intra- and intermolecular major groove contacts observed in the structure. In all calculated structures, Tyr-81 is stacked on the U10 bulge nucleotide, generating a hydrophobic pocket defined by Hδ1 and Hε1 of Tyr-81, the aliphatic portions of the Arg-70 and Arg-73 side chains, and the nonpolar regions of the H5/H6 edge of U10 in which the Ile-79 side chain resides (Fig. 4A). The hydrophobic nature of Ile-79 is an important determinant of binding specificity in the BIV complex (27). In addition to the extensive van der Waals contacts, the Tyr-81 hydroxyl group is positioned in all calculated structures to hydrogen bond to the Arg-70 ηNH2 proton on the opposite β strand (Fig. 4B), potentially helping to stabilize the peptide conformation. Furthermore, the imidazole ring of His-80 is positioned in most structures to hydrogen bond to the Arg-73 guanidinium group (Fig. 4C), potentially further stabilizing the peptide structure.
Fig. 4.
Interactions involving the C-terminal end of the JDV Tat ARM and effects of mutations on binding affinity. (A) View of the average minimized structure showing the stacking interaction between Tyr-81 and the U10 residue and the surrounding hydrophobic pocket delineated by the side chains of Arg-70, Arg-73, and Ile-79. (B) View showing the likely hydrogen bonding interactions of the Arg-70 (orange) guanidinium group with the Tyr-81 (blue) hydroxyl group and the Hoogsteen face of G11 located above the base triple (green). Groups within hydrogen bonding distance are indicated by dashed lines. (C) View of one representative structure showing the positions of His-80 and Arg-73 within hydrogen-bonding distance. The positions of the Arg-70, Arg-73, and His-80 side chains are well defined in the ensemble of structures despite the absence of electrostatic constraints imposed on the calculations. (D) Binding of the JDV Tat peptide and mutants to BIV TAR using gel mobility-shift assays. Binding reactions were performed at the peptide concentrations indicated with 0.2 nM RNA, and dissociation constants were calculated as described in Materials and Methods.(E) Binding of the JDV Tat peptide and Y81R mutant to BIV TAR and U10C mutant RNAs. For each peptide, relative association constants, as determined in D, were normalized to the values obtained with BIV TAR.
Costabilization of Peptide and RNA Structure Through an Intermolecular Interaction. Tyr-81 participates in a rather extensive network of interactions that includes crossstrand peptide interactions and stacking on the U10 bulge of the base triple, which is surprisingly stable in the JDV complex. To test whether Tyr-81 participates in simultaneously stabilizing the peptide and RNA structure and whether this contributes to the unusually high affinity of the JDV peptide–BIV TAR interaction, we measured RNA-binding affinities with a set of peptide and RNA mutants. Substituting Tyr-81 with Phe reduces affinity by ≈6-fold (Fig. 4D), consistent with a role for the hydroxyl group in hydrogen bonding to Arg-70 on the opposite strand that “locks” the peptide ends. Removing the aromatic ring by substitution with the Arg side chain of BIV Tat appears to reduce affinity even further (≈9-fold), as also observed previously (33), suggesting that Tyr-81 interactions within the hydrophobic pocket also contribute to the high affinity, perhaps also helping to stabilize the bound peptide conformation.
Although the stacking of Tyr-81 on the U10 bulged base appears to position Tyr to interact with the opposing β-strand, it also may help stabilize the bound TAR conformation by positioning U10 in the plane of the A13:U24 pair to form a base triple (Fig. 4B). This synergistic structural relationship is consistent with the measured binding affinities of the JDV peptide or Arg-81 mutant with a U10 bulge mutant RNA (Fig. 4E). Eliminating the base triple by mutating U10 to C decreases affinity of the JDV peptide by >5-fold but decreases affinity of the Arg-81 mutant by only ≈2-fold. Thus, the combination of the base triple and Tyr mutations is not significantly more detrimental than the Tyr mutation alone, supporting a structural view in which the peptide and RNA structures are stabilized in a highly interdependent manner, mediated largely by the Tyr-base stacking interaction. Aromatic amino acid stacking interactions have been found to stabilize other types of RNA structures, including a tetraloop in bacteriophage N peptide–RNA complexes and other loop structures in U1 hairpin and ribosomal RNA complexes (45–50), but these interactions are not also part of stabilizing networks within the protein, as in the JDV complex.
Induction of an HIV-1 TAR-Like Binding Site and Implications for Drug Design. As mentioned earlier, BIV TAR bound to the BIV Tat peptide and HIV TAR bound to argininamide or to the HIV Tat peptide adopt very similar conformations, except for the positioning of the U10 bulge base (analogous to U23 in HIV; Fig. 1C). However, when BIV TAR is bound to the JDV peptide, the base triple becomes stabilized, primarily via the Tyr-81–U10 stacking interaction, and even the bulge base becomes virtually superimposable on the bound HIV-1 TAR structure (Fig. 5). Thus, the additional protein–RNA interactions observed in the JDV chameleon peptide, largely involving just one amino acid different from BIV Tat (Tyr-81 versus Arg-81), may represent another evolutionary pathway to evolve an RNA-binding peptide with multiple specificities, in this case via a structural costabilization mechanism. Although the JDV chameleon does bind HIV TAR, it does not use the β-hairpin binding mode (33), perhaps because the bulge region of HIV TAR in its unbound state is more disordered than unbound BIV TAR. Nonetheless, the striking structural similarities between HIV and BIV TAR are intriguing and suggest that other peptide variants might evolve to compensate for differences in RNA structure. Attempts have been made to create HIV TAR binders by constraining the BIV Tat β-hairpin to minimize the energetic cost of peptide folding, and these show somewhat enhanced affinities for HIV TAR (51, 52). It may be desirable to also include C-terminal residues of the JDV Tat ARM to further validate the costabilization hypothesis and to develop even tighter binders. The critical importance of a single arginine contact to HIV TAR (19, 20) also raises the possibility that small molecules incorporating both guanidinium and aromatic moieties might be used to mimic the Tat contact while simultaneously stabilizing the bound RNA structure.
Fig. 5.
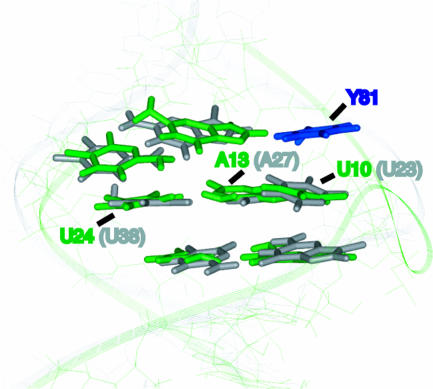
Superposition of the binding regions of BIV TAR complexed to the JDV ARM (green) and HIV-1 TAR complexed to argininamide (gray). Bases important for the binding of ARM peptides to BIV TAR (thick bases) (26) were used to overlap the JDV complex with an NMR model of the HIV-1 TAR complex (19). In the JDV complex, Tyr-81 (blue) stacks on the U10 bulge base of BIV TAR, positioning it to hydrogen bond to the A13:U24 base pair and form the base triple.
Conclusions
The structure of the JDV peptide–BIV TAR complex shows how the affinity of an RNA–protein interaction can be increased not only by the addition of intermolecular contacts but also by stabilizing the structures of the two binding partners, in this case resulting largely from one stacking interaction between Tyr-81 and the bulged U10 base. The study reinforces the importance of induced fit in RNA–protein recognition and highlights one type of cofolding mechanism that can occur between partially disordered segments of protein and RNA, probably driven by the hydrophobic nature of the interaction.
One intriguing observation is the resemblance between the bound structure of HIV TAR and BIV TAR bound to the JDV peptide, further supporting the view that there should be no fundamental barrier to accommodating a β-hairpin peptide in the HIV TAR-binding site (30). Peptides or small molecules that bind HIV TAR with high affinity may function as HIV inhibitors given the critical role of the Tat–TAR interaction in viral replication, and the contacts seen in the JDV complex suggest additional types of interactions that may target HIV TAR, particularly by using an aromatic moiety to help stabilize the base triple. Clearly, it will be interesting to characterize the structures of tight binding chameleon peptides or other variants and to assess their effectiveness as HIV inhibitors.
Supplementary Material
Acknowledgments
We thank Karim Dabbagh, Anwer Mujeeb, Nick Ulyanov, and members of the Frankel laboratory for helpful suggestions, and Chandreyee Das, Nick Ulyanov, and John Gross for comments on the manuscript. This work was supported by a Howard Hughes Medical Institutes predoctoral fellowship (to M.D.D.) and by National Institutes of Health Grants GM56531 and AI29135 (to A.D.F.).
Author contributions: A.D.F. designed research; V.C. and M.D.D. performed research; V.C. and M.D.D. analyzed data; and V.C. and A.D.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ARM, arginine-rich motif; BIV, bovine immunodeficiency virus; JDV, Jembrana disease virus.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1ZBN).
References
- 1.Frankel, A. D. & Smith, C. A. (1998) Cell 92, 149–151. [DOI] [PubMed] [Google Scholar]
- 2.Sundberg, E. J. & Mariuzza, R. A. (2000) Struct. Fold. Des. 8, R137–R142. [DOI] [PubMed] [Google Scholar]
- 3.Dyson, H. J. & Wright, P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60. [DOI] [PubMed] [Google Scholar]
- 4.Goh, C. S., Milburn, D. & Gerstein, M. (2004) Curr. Opin. Struct. Biol. 14, 104–109. [DOI] [PubMed] [Google Scholar]
- 5.Abdul-Manan, N., Aghazadeh, B., Liu, G. A., Majumdar, A., Ouerfelli, O., Siminovitch, K. A. & Rosen, M. K. (1999) Nature 399, 379–383. [DOI] [PubMed] [Google Scholar]
- 6.Kim, A. S., Kakalis, L. T., Abdul-Manan, N., Liu, G. A. & Rosen, M. K. (2000) Nature 404, 151–158. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez, R., Salazar, G., Baldridge, K. K. & Romesberg, F. E. (2003) Proc. Natl. Acad. Sci. USA 100, 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James, L. C., Roversi, P. & Tawfik, D. S. (2003) Science 299, 1362–1367. [DOI] [PubMed] [Google Scholar]
- 9.Williamson, J. R. (2003) RNA 9, 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayana, N. & Weiss, M. A. (1999) Biopolymers 48, 167–180. [DOI] [PubMed] [Google Scholar]
- 11.Frankel, A. D. (2000) Curr. Opin. Struc. Biol. 10, 332–340. [DOI] [PubMed] [Google Scholar]
- 12.Hermann, T. & Patel, D. J. (2000) Science 287, 820–825. [DOI] [PubMed] [Google Scholar]
- 13.Weeks, K. M., Ampe, C., Schultz, S. C., Steitz, T. A. & Crothers, D. M. (1990) Science 249, 1281–1285. [DOI] [PubMed] [Google Scholar]
- 14.Weeks, K. M. & Crothers, D. M. (1991) Cell 66, 577–588. [DOI] [PubMed] [Google Scholar]
- 15.Calnan, B. J., Tidor, B., Biancalana, S., Hudson, D. & Frankel, A. D. (1991) Science 252, 1167–1171. [DOI] [PubMed] [Google Scholar]
- 16.Tao, J. & Frankel, A. D. (1992) Proc. Natl. Acad. Sci. USA 89, 2723–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao, J. & Frankel, A. D. (1993) Proc. Natl. Acad. Sci. USA 90, 1571–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long, K. S. & Crothers, D. M. (1995) Biochemistry 34, 8885–8895. [DOI] [PubMed] [Google Scholar]
- 19.Puglisi, J. D., Tan, R., Calnan, B. J., Frankel, A. D. & Williamson, J. R. (1992) Science 257, 76–80. [DOI] [PubMed] [Google Scholar]
- 20.Aboul-ela, F., Karn, J. & Varani, G. (1995) J. Mol. Biol. 253, 313–332. [DOI] [PubMed] [Google Scholar]
- 21.Long, K. S. & Crothers, D. M. (1999) Biochemistry 38, 10059–10069. [DOI] [PubMed] [Google Scholar]
- 22.Dayie, K. T., Brodsky, A. S. & Williamson, J. R. (2002) J. Mol. Biol. 317, 263–278. [DOI] [PubMed] [Google Scholar]
- 23.Pitt, S. W., Majumdar, A., Serganov, A., Patel, D. J. & Al-Hashimi, H. M. (2004) J. Mol. Biol. 338, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter, S., Nadin-Davis, S. A., Wannemuehler, Y. & Roth, J. A. (1993) J. Virol. 67, 4399–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garvey, K. J., Oberste, M. S., Elser, J. E., Braun, M. J. & Gonda, M. A. (1990) Virology 175, 391–409. [DOI] [PubMed] [Google Scholar]
- 26.Chen, L. & Frankel, A. D. (1994) Biochemistry 33, 2708–2715. [DOI] [PubMed] [Google Scholar]
- 27.Chen, L. & Frankel, A. D. (1995) Proc. Natl. Acad. Sci. USA 92, 5077–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puglisi, J. D., Chen, L., Blanchard, S. & Frankel, A. D. (1995) Science 270, 1200–1203. [DOI] [PubMed] [Google Scholar]
- 29.Ye, X., Kumar, R. A. & Patel, D. J. (1995) Chem. Biol. 2, 827–840. [DOI] [PubMed] [Google Scholar]
- 30.Smith, C. A., Crotty, S., Harada, Y. & Frankel, A. D. (1998) Biochemistry 37, 10808–10814. [DOI] [PubMed] [Google Scholar]
- 31.Chen, H., He, J., Fong, S., Wilcox, G. & Wood, C. (2000) J. Virol. 74, 2703–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chadwick, B. J., Coelen, R. J., Wilcox, G. E., Sammels, L. M. & Kertayadnya, G. (1995) J. Gen. Virol. 76, 1637–1650. [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. A., Calabro, V. & Frankel, A. D. (2000) Mol. Cell 6, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 34.Milligan, J. F. & Uhlenbeck, O. C. (1989) Methods Enzymol. 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 35.Kao, C., Zheng, M. & Rudisser, S. (1999) RNA 5, 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smallcombe, S. H. (1993) J. Am. Chem. Soc. 115, 4776–4785. [Google Scholar]
- 37.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 227–293. [DOI] [PubMed] [Google Scholar]
- 38.Goddard, T. & Kneller, D. G. (1998) sparky 3 (University of California, San Francisco).
- 39.Wutrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 40.Guntert, P., Mumenthaler, C. & Wuthrich, K. (1997) J. Mol. Biol. 273, 283–298. [DOI] [PubMed] [Google Scholar]
- 41.Wuthrich, K., Billeter, M. & Braun, W. (1983) J. Mol. Biol. 169, 949–961. [DOI] [PubMed] [Google Scholar]
- 42.Saenger, W. (1984) Principles of Nucleic Acid Structure (Springer, New York).
- 43.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 44.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 45.Legault, P., Li, J., Mogridge, J., Kay, L. E. & Greenblatt, J. (1998) Cell 93, 289–299. [DOI] [PubMed] [Google Scholar]
- 46.Cai, Z., Gorin, A., Frederick, R., Ye, X., Hu, W., Majumdar, A., Kettani, A. & Patel, D. J. (1998) Nat. Struct. Biol. 5, 203–212. [DOI] [PubMed] [Google Scholar]
- 47.Avis, J. M., Allain, F. H., Howe, P. W., Varani, G., Nagai, K. & Neuhaus, D. (1996) J. Mol. Biol. 257, 398–411. [DOI] [PubMed] [Google Scholar]
- 48.Blakaj, D. M., McConnell, K. J., Beveridge, D. L. & Baranger, A. M. (2001) J. Am. Chem. Soc. 123, 2548–2551. [DOI] [PubMed] [Google Scholar]
- 49.Shiels, J. C., Tuite, J. B., Nolan, S. J. & Baranger, A. M. (2002) Nucleic Acids Res. 30, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao, H., White, S. A. & Williamson, J. R. (1999) Nat. Struct. Biol. 6, 1139–1147. [DOI] [PubMed] [Google Scholar]
- 51.Runyon, S. T. & Puglisi, J. D. (2003) J. Am. Chem. Soc. 125, 15704–15705. [DOI] [PubMed] [Google Scholar]
- 52.Athanassiou, Z., Dias, R. L. A., Moehle, K., Dobson, N., Varani, G. & Robinson, J. A. (2004) J. Am. Chem. Soc. 126, 6906–6913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.