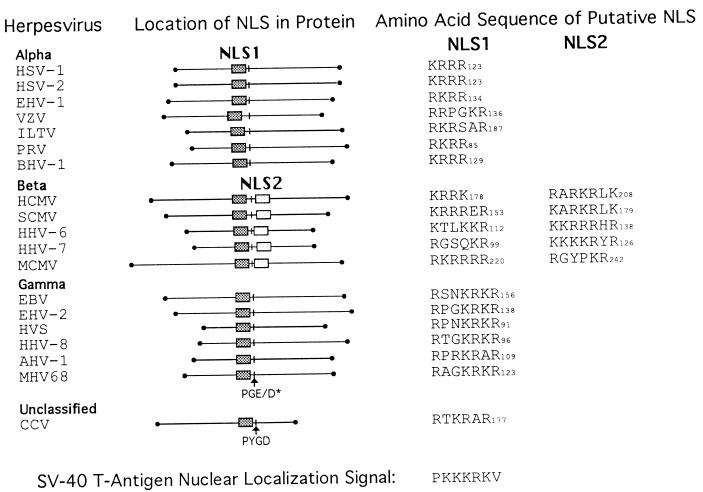
FIG. 11.
Positional homologs of NLS1 and NLS2 in the pAP homologs of herpes-group viruses. Amino acid sequences of available pAP homologs were aligned by using the Pima program (Baylor College of Medicine), and positional homologs of CMV pAP NLS1 and NLS2 were identified as indicated. A conserved Pro-Gly-Glu/Asp (PGE/D) sequence is indicated as an internal reference, and the carboxy-terminal amino acid number for each sequence is given. Sequences were obtained from GenBank; the viruses are as follows: HSV-1 (40), HSV-2 (54), equine herpesvirus 1 (EHV-1) (57), varicella-zoster virus (VZV) (12), infectious laryngotracheitis virus (ILTV) (27), pseudorabies virus (PRV) (7), bovine herpesvirus 1 (BHV-1) (28), HCMV strain AD169 (9), SCMV strain Colburn (60), human herpesvirus 6 (HHV-6) (26), human herpesvirus 7 (HHV-7) (41), murine CMV (MCMV) (39), Epstein-Barr virus (EBV) (2), equine herpesvirus 2 (EHV-2) (56), herpesvirus saimiri (HVS) (1), human herpesvirus 8 (HHV-8) (51), wildebeest herpesvirus (AHV-1) (17), murine herpesvirus 68 (MHV68) (59), and channel catfish virus (CCV) (11). Classification as alpha-, beta-, or gammaherpesvirus is based on the nomenclature of Roizman et al. (50). The amino acid sequence of the SV40 large-T-antigen NLS (33, 34) is shown at the bottom.