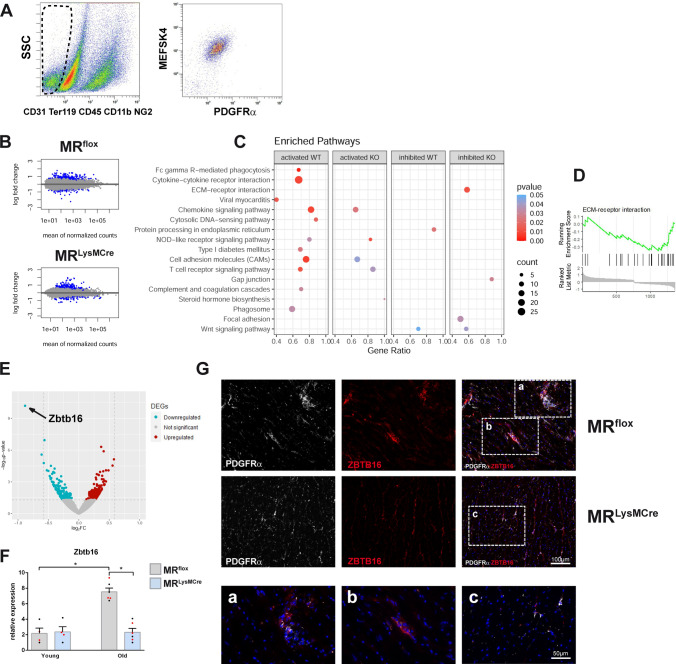
Fig. 2.
Macrophage MR deficiency affects the transcriptome of fibroblasts in the aging heart. A Gating strategy to identify CD31−/TER119−/CD45−/CD11b−/NG2−/MEFSK4+/PDGFRα+ cells. Fibroblasts were isolated by flow cytometry from hearts of male/female young and old MRflox and MRLysMcre mice. B MA plots of gene expression in fibroblasts. Significantly differentially expressed genes (old vs. young) with adjusted p values < 0.1 are indicated in blue. C Graphical dot plot representation of the most significantly enriched pathways in fibroblasts. Comparison of altered pathways (p < 0.01) between MRflox (WT) and MRLysMCre (KO) mice determined by R package clusterProfiler. Large gene ratio in combination with high count and low p value indicates a more pronounced activation or inhibition for a specific pathway; D GSEA plot of downregulated ECM–receptor interaction pathway in fibroblasts from MRLysMCre mice. E Volcano plot of contrast of old cardiac fibroblasts from MRLysMcre vs. MRflox mice. F Relative expression of Zbtb16 in young/old fibroblasts from MRflox and MRLysMCre hearts. Depicted points in red/black indicate, respectively, male and female mice. G Immunofluorescence micrographs of heart sections from old MRflox and MRLysMCre mice showing fibroblasts (PDGFRα+ cells) and ZBTB16 immunoreactivity. MRLysMCre hearts displayed a weak expression of ZBTB16 in PDGFRα-positive fibroblasts. Nuclei were stained with NucBlue™. Mean ± SEM, n = 4–6 per group; *p < 0.05