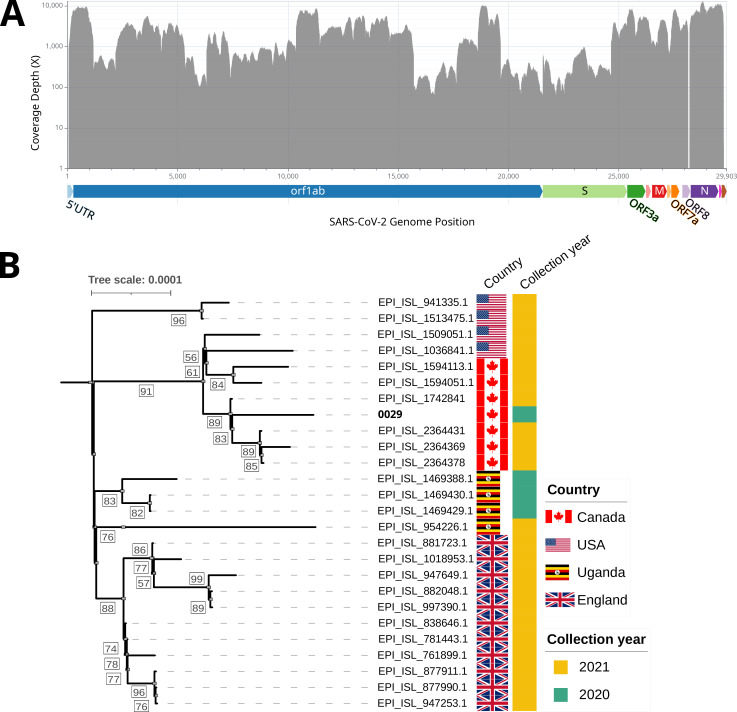
Fig 1.
(A) A barplot of the sequencing coverage depth across the SARS-CoV-2 genome of the lineage A.23.1 sequence recovered from a Canadian cat (sample WIN-AH-2021-OTH-Kari-0029-OS-1) generated using wgscovplot (https://github.com/nhhaidee/wgscovplot). The x-axis shows the SARS-CoV2 genome position, and the y-axis shows genome coverage depth. At the bottom, the whole genome of the SARS-CoV2 reference strain, including gene features, is attached. (B) A maximum-likelihood phylogenetic tree using the whole genome of lineage A.23.1 SARS-CoV-2 sequence from a Canadian cat (sample WIN-AH-2021-OTH-Kari-0029-OS-1; denoted as 0029 in the tree) along with 25 most closely related lineage A.23.1 sequences from GISAID (20) as identified by UShER phylogenetic placement analysis (2023-12-07) which are collected from different geographic regions but at the similar time. Sequence alignment was performed using MAFFT (v7.511) under the default settings (Method- FFT-NS-2) (21), and the maximum-likelihood phylogenetic tree was inferred using IQ-TREE (v1.6.12) with the K3Pu + F model (determined by IQ-TREE’s ModelFinder) and 1,000 ultra-fast bootstraps (22–24). SARS-CoV-2 Wuhan-Hu-1 reference sequence (MN908947.3) has been used as the outgroup.