Extended Data Fig. 5 |. Substrate-Binding in SLC4 and SLC26 Transporters.
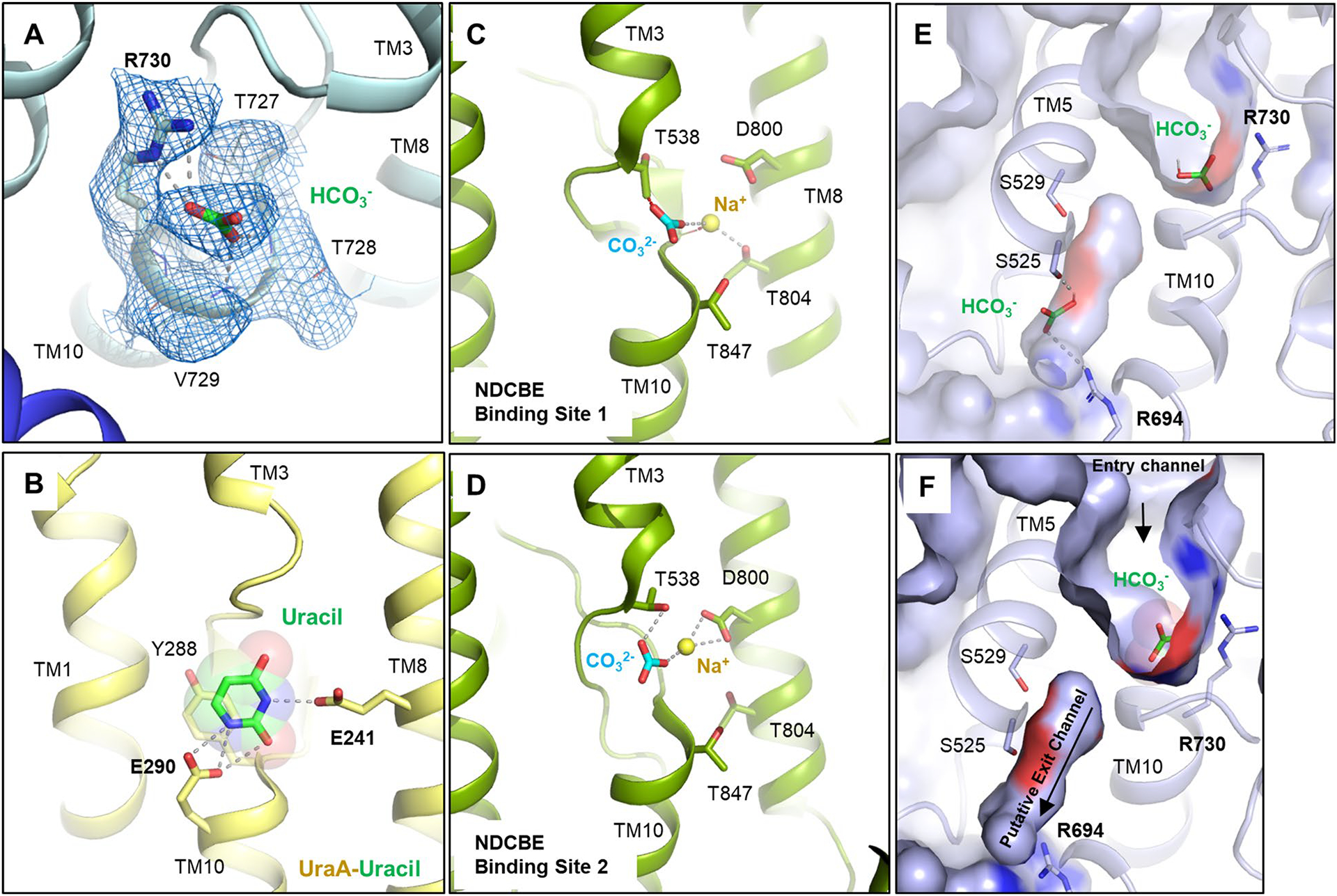
a, Cryo-EM density of bicarbonate and binding site residues (marine mesh) using a contour level of 6σ. b, Binding site of uracil in the UraA-Uracil structure (PDB ID: 3QE7)24. c, d, Carbonate and sodium ions in the rabbit NDCBE (SLC4A8) cryo-EM structure (PDB ID: 7RTM, EMD-24683)15, with clashes between CO32− and T538, as well as Na+ and the backbone shown as red dotted lines. Ionic interactions and hydrogen bonds are shown as grey dotted lines. e, SACP simulations allow to calculate bicarbonate affinity to AE1 anion binding site (see methods). In some structures (~5%) we have identified a second bicarbonate binding site in the putative exit channel. f, Surface display shows putative anion exit channel leading from the bicarbonate binding site to the cytoplasmic site. Channel extends below the plane of section.
