Extended Data Fig. 2 |. Cryo-EM Structure Determination of Apo AE1 and AE1-Bicarbonate complexes.
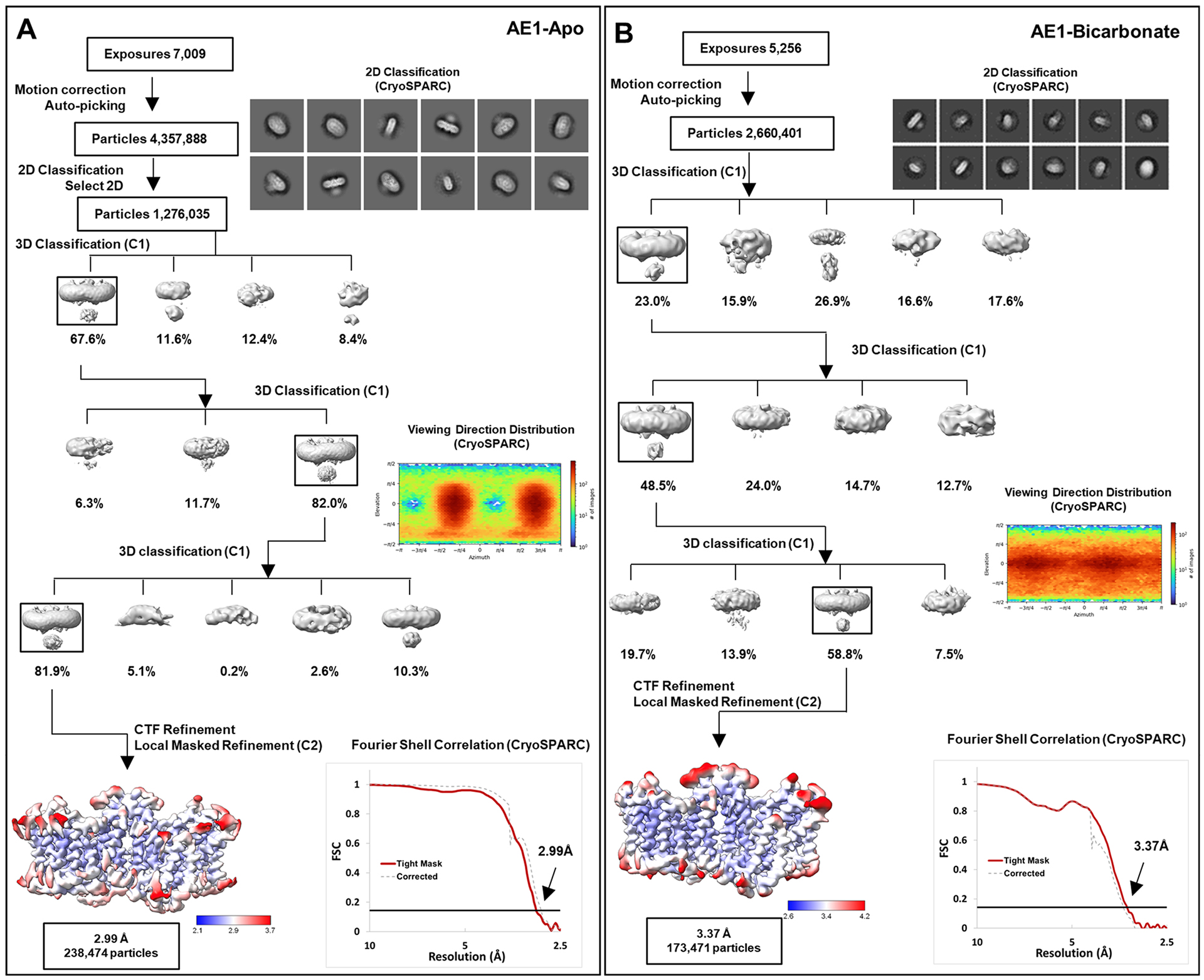
a, b, workflow of Apo AE1 (a) and AE1-Bicarbonate (b) data processing, showing representative 2D classes, followed by 2D classification, ab initio modeling and several rounds of 3D classification, with representative 3D classes shown. After multiple rounds of 3D classification, final particle stacks were subjected to CTF refinement followed by local refinement of the masked AE1 membrane domain with imposed C2 symmetry. GS-FSC calculations indicate global resolutions of 2.99 Å (Apo AE1) and 3.37 (AE1-Bicarbonate) applying the 0.143 cutoff. Viewing direction distribution analysis (cryoSPARC) indicates sufficient coverage. Initial models were built in PHENIX, and then further refined in ServalCat for the generation of final maps and coordinates. Calculations in cryoSPARC indicate local resolutions of lower than 3 Å around substrate and inhibitor binding sites.
