Fig. 1 |. The nonsense-mediated RNA decay pathway.
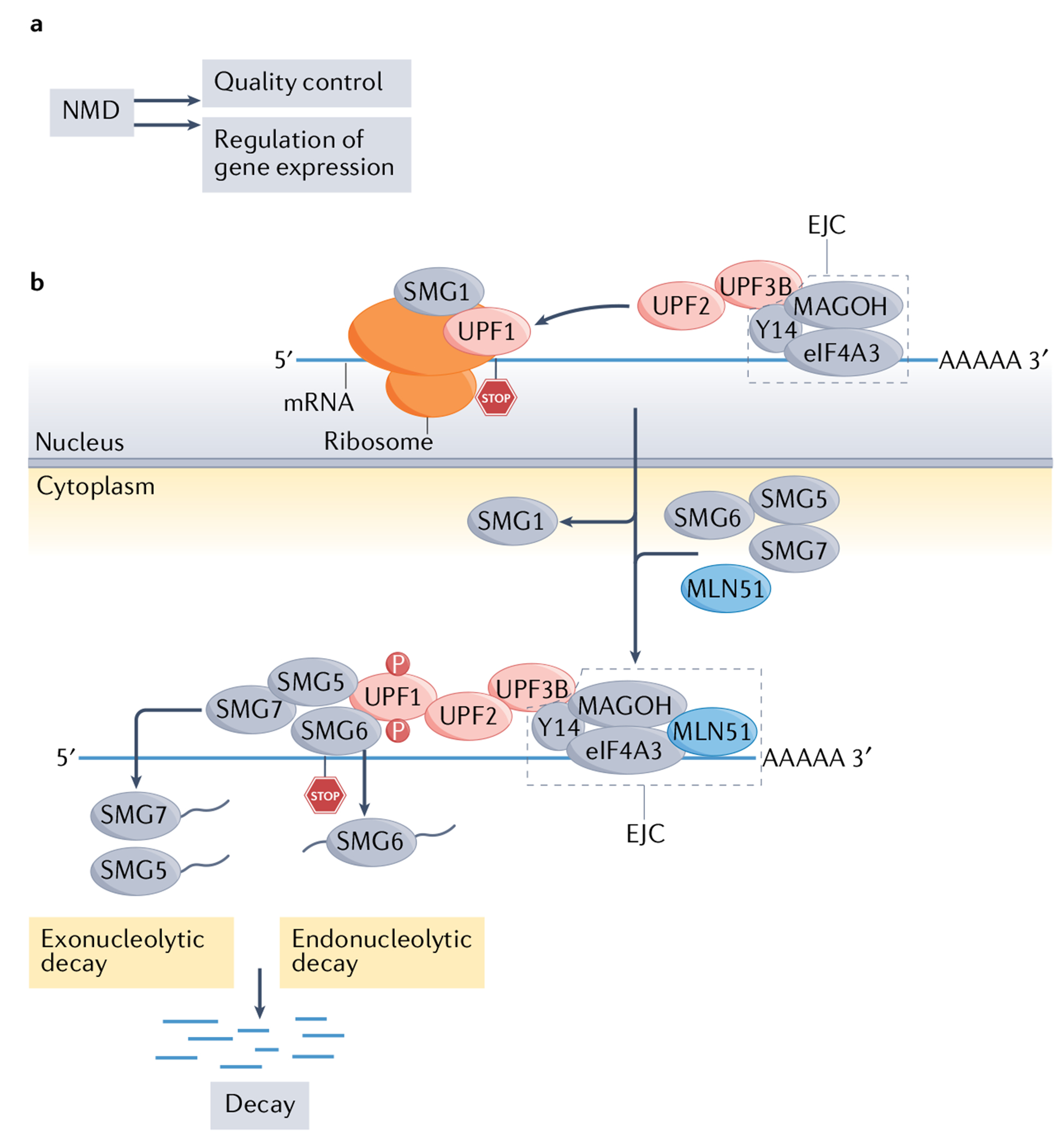
a | The two broad functions of the nonsense-mediated RNA decay (NMD) pathway. b | Simplified view of the molecular mechanism of NMD in mammals. Up-frameshift 1 (UPF1) is an RNA helicase recruited to all mRNAs that remains stably bound only to NMD target mRNAs by an unknown mechanism205; thus, high UPF1 occupancy is regarded as a reliable feature of NMD target mRNAs18. UPF1 also binds to UPF2, which triggers the former to undergo a conformational change that activates its RNA-helicase activity206. UPF2 also binds to UPF3B, which, in turn, interacts with the exon-junction complex (EJC)207, a set of proteins (consisting of eukaryotic translation initiation factor 4A-III (eIF4A3), Y14 (also known as RBM8A) and MAGOH45) that bind near exon–exon junctions in RNAs208 after they are spliced in the nucleus. EJC-bound RNAs are then transported to the cytoplasm, where accessory proteins, including the EJC protein MLN51 (also known as CASC3) are recruited. The formation of an EJC-core NMD factor complex triggers RNA decay, but other mechanisms can also elicit NMD (see FIG. 2). During this chain of events, the protein kinase SMG1 phosphorylates UPF1, which is thought to commit an mRNA to degradation, since phosphorylated UPF1 binds to the RNA endonuclease SMG6, which is known to cleave NMD target mRNAs18. Phosphorylated UPF1 also recruits the SMG5–SMG7 heterodimer104, which interacts with decapping and deadenylating enzymes to further foster RNA decay. NMD is regulated by many factors, including eIF2α phosphorylation during cellular stress, microRNAs and specific regulatory proteins (not shown)18,165,166.
